An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments - PubMed (original) (raw)
Comparative Study
An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments
Colin Echeverría Aitken et al. Biophys J. 2008.
Abstract
The application of single-molecule fluorescence techniques to complex biological systems places demands on the performance of single fluorophores. We present an enzymatic oxygen scavenging system for improved dye stability in single-molecule experiments. We compared the previously described protocatechuic acid/protocatechuate-3,4-dioxygenase system to the currently employed glucose oxidase/catalase system. Under standardized conditions, we observed lower dissolved oxygen concentrations with the protocatechuic acid/protocatechuate-3,4-dioxygenase system. Furthermore, we observed increased initial lifetimes of single Cy3, Cy5, and Alexa488 fluorophores. We further tested the effects of chemical additives in this system. We found that biological reducing agents increase both the frequency and duration of blinking events of Cy5, an effect that scales with reducing potential. We observed increased stability of Cy3 and Alexa488 in the presence of the antioxidants ascorbic acid and n-propyl gallate. This new O(2)-scavenging system should have wide application for single-molecule fluorescence experiments.
Figures
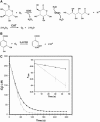
Figure 1
Comparison of GODCAT and PCD O2-scavenging systems. (A and B) Schematics of chemical reactions in GODCAT and PCD systems, respectively. (C) Dissolved oxygen time courses for GODCAT (solid) and standardized PCD (shaded) systems. Conditions for each reaction are as follows: GODCAT ∼100 nM glucose oxidase, ∼1.5 _μ_M catalase, 56 mM glucose; PCD ∼50 nM PCD, 2.5 mM PCA. (C inset) Time courses of PCA consumption as monitored by absorbance at 290 nm. Each reaction contains 200 _μ_M PCA and either 50 nM (solid) or 10 nM (shaded) PCD.
Figure 2
Analysis of single fluorophores. (A) Representative color overlay of all fluorophores observed in one acquisition movie. (B) Representative fluorescence versus time trace for a single dye. Initial events (1), off-blink (2), and on-blink events (3) were segregated before lifetime analysis; events limited by acquisition length (4) were discarded. (C) Initial, off-blink, and on-blink event distributions were fit, as described, to an exponential probability function to determine mean lifetimes, here Cy5 initial lifetimes with PCD.
Figure 3
Characterization of dye stability in the GODCAT and PCD systems. (A) Table of mean lifetimes and event frequencies (per molecule) in the GODCAT system. (B) Table of mean lifetimes and event frequencies (per molecule) in the standardized PCD system. (C) Comparison of initial dye lifetimes in the standardized PCD (open), dilute PCD (light shaded), GODCAT (shaded), and GODCAT+PCA (dark shaded) systems.
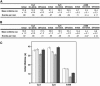
Figure 4
Cy5 dye stability in the presence of biological reducing agents. (A) Table of event frequencies and lifetimes for initial, on-blink, and off-blink events (per molecule) in the absence or presence of 10 mM BME, DTT, and TCEP. Lifetime distributions are fit as described in Materials and Methods. (B) Bar plot of the ratios of on-blink/initial lifetimes (open) and off-blink/initial lifetimes (shaded) for Cy5 in the presence of reducing agents. (C) Bar plot of the signal/noise ratio for Cy5, determined on a per molecule basis.
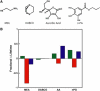
Figure 5
Effects of additives on initial lifetimes of Cy3, Cy5, and Alexa488. (A) Chemical structures of additives: MEA, DABCO, ascorbic acid, and nPG. (B) Bar plot of the fractional change in initial lifetimes of Cy3 (green), Cy5 (red), and Alexa488 (blue) in the PCD system with respective additives.
Similar articles
- Absence of nuclease activity in commonly used oxygen-scavenging systems.
Gahlon HL, Poudevigne-Durance P, Rueda D. Gahlon HL, et al. BMC Res Notes. 2017 Nov 21;10(1):606. doi: 10.1186/s13104-017-2929-6. BMC Res Notes. 2017. PMID: 29162131 Free PMC article. - An oxygen-scavenging system without impact on DNA mechanical properties in single-molecule fluorescence experiments.
Gao S, Liang J, Tan C, Ma J. Gao S, et al. Nanoscale. 2025 Feb 6;17(6):3236-3242. doi: 10.1039/d4nr04287e. Nanoscale. 2025. PMID: 39633609 - Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments.
Swoboda M, Henig J, Cheng HM, Brugger D, Haltrich D, Plumeré N, Schlierf M. Swoboda M, et al. ACS Nano. 2012 Jul 24;6(7):6364-9. doi: 10.1021/nn301895c. Epub 2012 Jun 22. ACS Nano. 2012. PMID: 22703450 Free PMC article. - Fluorescence microscopy for visualizing single-molecule protein dynamics.
Yokota H. Yokota H. Biochim Biophys Acta Gen Subj. 2020 Feb;1864(2):129362. doi: 10.1016/j.bbagen.2019.05.005. Epub 2019 May 10. Biochim Biophys Acta Gen Subj. 2020. PMID: 31078674 Review. - Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments.
Levitus M, Ranjit S. Levitus M, et al. Q Rev Biophys. 2011 Feb;44(1):123-51. doi: 10.1017/S0033583510000247. Epub 2010 Nov 26. Q Rev Biophys. 2011. PMID: 21108866 Review.
Cited by
- Membrane binding controls ordered self-assembly of animal septins.
Szuba A, Bano F, Castro-Linares G, Iv F, Mavrakis M, Richter RP, Bertin A, Koenderink GH. Szuba A, et al. Elife. 2021 Apr 13;10:e63349. doi: 10.7554/eLife.63349. Elife. 2021. PMID: 33847563 Free PMC article. - Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging.
Ha T, Tinnefeld P. Ha T, et al. Annu Rev Phys Chem. 2012;63:595-617. doi: 10.1146/annurev-physchem-032210-103340. Epub 2012 Jan 30. Annu Rev Phys Chem. 2012. PMID: 22404588 Free PMC article. Review. - Accelerated single photon emission from dye molecule-driven nanoantennas assembled on DNA.
Busson MP, Rolly B, Stout B, Bonod N, Bidault S. Busson MP, et al. Nat Commun. 2012 Jul 17;3:962. doi: 10.1038/ncomms1964. Nat Commun. 2012. PMID: 22805569 - Single molecule targeted sequencing for cancer gene mutation detection.
Gao Y, Deng L, Yan Q, Gao Y, Wu Z, Cai J, Ji D, Li G, Wu P, Jin H, Zhao L, Liu S, Ge L, Deem MW, He J. Gao Y, et al. Sci Rep. 2016 May 19;6:26110. doi: 10.1038/srep26110. Sci Rep. 2016. PMID: 27193446 Free PMC article. - Thermodynamic origins of monovalent facilitated RNA folding.
Holmstrom ED, Fiore JL, Nesbitt DJ. Holmstrom ED, et al. Biochemistry. 2012 May 8;51(18):3732-43. doi: 10.1021/bi201420a. Epub 2012 Apr 23. Biochemistry. 2012. PMID: 22448852 Free PMC article.
References
- Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. - PubMed
- Cornish P.V., Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007;2:53–61. - PubMed
- Zhuang X., Rief M. Single-molecule folding. Curr. Opin. Struct. Biol. 2003;13:88–97. - PubMed
- Schuler B. Single-molecule fluorescence spectroscopy of protein folding. ChemPhysChem. 2005;6:1206–1220. - PubMed
- Lipman E.A., Schuler B., Bakajin O., Eaton W.A. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources