Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes - PubMed (original) (raw)
Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes
Gregory D Gregory et al. Mol Cell Biol. 2007 Dec.
Abstract
Histone lysine methylation regulates genomic functions, including gene transcription. Previous reports found various degrees of methylation at H3K4, H3K9, and H4K20 within the transcribed region of active mammalian genes. To identify the enzymes responsible for placing these modifications, we examined ASH1L, the mammalian homolog of the Drosophila melanogaster Trithorax group (TrxG) protein Ash1. Drosophila Ash1 has been reported to methylate H3K4, H3K9, and H4K20 at its target sites. Here we demonstrate that mammalian ASH1L associates with the transcribed region of all active genes examined, including Hox genes. The distribution of ASH1L in transcribed chromatin strongly resembles that of methylated H3K4 but not that of H3K9 or H4K20. Accordingly, the SET domain of ASH1L methylates H3K4 in vitro, and knockdown of ASH1L expression reduced H3K4 trimethylation at HoxA10 in vivo. Notably, prior methylation at H3K9 reduced ASH1L-mediated methylation at H3K4, suggesting cross-regulation among these marks. Drosophila ash1 and trithorax interact genetically, and the mammalian TrxG protein MLL1 and ASH1L display highly similar distributions and substrate specificities. However, by using MLL null cell lines we found that their recruitments occur independently of each other. Collectively, our data suggest that ASH1L occupies most, if not all, active genes and methylates histone H3 in a nonredundant fashion at a subset of genes.
Figures
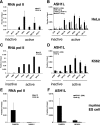
FIG. 1.
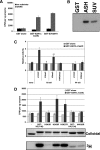
ASH1L is associated with actively transcribed genes. (A) ChIP analysis, with HeLa cells, of RNA Pol II at the active genes RPLP0, PPIA, RPS2, EEF1A1, and RPL41 and the inactive genes CD4 and MYT1. (B) ChIP assay using anti-ASH1L antibodies 296 and ab4477 at the genes listed above. (C and D) Similar analyses were performed with undifferentiated K562 cells. (E and F) RNA Pol II and ASH1L occupancy at the inactive NKX2.2 and active PABPC1 genes in murine embryonic stem (ES) cells. Error bars represent standard deviations. Highly transcribed and ubiquitously expressed active genes were identified using the RNA expression database SymAtlas, v1.2.4, which is maintained by the Genomics Institute of the Novartis Research Foundation (
http://symatlas.gnf.org/SymAtlas/
) (see Fig. S1A in the supplemental material) (50). rab Ig, rabbit immunoglobulin G.
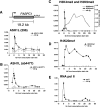
FIG. 2.
Histone methylation and ASH1L occupancy at the PABPC1 gene. ChIP assays of HeLa cells with antibodies against (A) ASH1L (296), (B) ASH1L (ab4477), (C) H3K4me3 and H3K9me3, (D) H4K20me1, and (E) RNA Pol II. All error bars represent standard deviations. Anti-ASH1L ChIP results are the averages of three to five independent experiments per primer pair. Nonimmune isotype-matched rabbit immunoglobulin G (rab Ig) was used as the control. ChIP assays using a third antibody specific for ASH1L (337ap) are shown in Fig. S1B in the supplemental material.
FIG. 3.
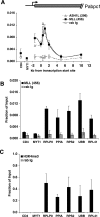
Recruitment of ASH1L correlates with onset of gene expression. (A) Diagram of the murine β-globin gene locus (not drawn to scale). ChIP analysis of (B) RNA Pol II, (C) ASH1L (antibody ab4477), and (D) H3K4me3 in murine G1E cells before or after differentiation with estradiol for 21 h. Error bars represent standard deviations for three independent experiments. HS, DNase 1 hypersensitive site; IVR, intervening region; TR, transcribed region; rab Ig, rabbit immunoglobulin G.
FIG. 4.
ASH1L occupancy persists following a transcription elongation block. ChIP with RNA Pol II, H3K4me3, and ASH1L (ab4477) antibodies at indicated sites at the PABPC1 gene in HeLa cells with (+ DRB) or without (no DRB) treatment with DRB for 6 h. Data were plotted as severalfold changes from levels for nontreated cells. Error bars represent standard deviations. Similar analyses were performed with G1E cells and are shown as Fig. S3 in the supplemental material.
FIG. 5.
The CsetC domain of ASH1L methylates H3K4 in vitro and is partially inhibited by preexisting H3K9 trimethylation. (A) HMTase activity of the SET domain of ASH1L (GST-ASH1L-CsetC), SUV39H1 (GST-SUV39H1), or GST alone with OGNS as the substrate. Four microliters of assembled OGNS, equating to a total of 108 ng of core histones, was used per 20-μl reaction. (B) HMTase assays with OGNS as the substrate were scaled up (6×), resolved on a 10% SDS-polyacrylamide gel, and treated with Autofluor before exposure to film for 9 days. (C) HMTase activity of the SET domain of ASH1L (GST-ASH1L-CsetC) or GST with histone H3 and H4 peptides as substrates. The value obtained with GST alone was set to 1. Premethylation of H3K4 completely blocks activity, whereas H3K9 premethylation partially inhibits activity. *, P < 0.001; **, P < 0.05; unmod, unmodified. (D) HMTase assay using GST-H3(1-46) and GST-H4(1-34) with indicated lysine-to-arginine substitutions as substrates. Graphs represent scintillation counts from filter assays. Error bars denote standard deviations (top). Part of the reaction mixture was analyzed by SDS-polyacrylamide gel electrophoresis and Colloidal blue staining (middle) prior to autoradiography (bottom).
FIG. 6.
Overlapping chromatin occupancies of ASH1L and MLL1 at housekeeping genes. (A) ChIP with anti-ASH1L (296), anti-MLL1 (antibody 456), and control rabbit immunoglobulin G (rab Ig) at PABPC1 in 293T cells. (B and C) ChIP analysis, with 293T cells, of MLL1 (456) (B) and H3K4me3 (C) at the active genes RPLP0, PPIA, RPS2, UBB, and RPL41 and the inactive genes CD4 and MYT1. Error bars represent standard deviations.
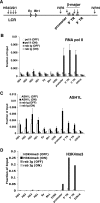
FIG. 7.
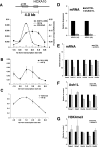
ASH1L is required for full Hox gene expression and H3K4 trimethylation. (A) ChIP with anti-H3K4me3 and anti-ASH1L (296) antibodies at the active HoxA10 gene. (B) ChIP analysis of MLL1 (456) across the HoxA10 gene. (C) ChIP with anti-H3K4me2 across the HoxA10 gene. Error bars represent standard deviations. rab Ig, rabbit immunoglobulin G. (D) Efficiency of knockdown of ASH1L mRNA, determined using primers directed towards the middle (#1) and 3′ (#2) regions of ASH1L cDNA. (E) mRNA levels of indicated genes, as determined by quantitative real-time RT-PCR with shCTRL and shASH1L 293T cells. (F) ASH1L occupancy and (G) H3K4me3 levels at +0.5 to 1.0 kb of indicated genes in shCTRL and shASH1L 293T cells.
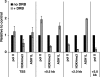
FIG. 8.
ASH1L occupancy is independent of MLL1. (A) Real-time RT-PCR analysis of HoxA9, Meis1, and TubA2 mRNA expression in Mll1+/+ and _Mll1_−/− MEFs. (B and C) ChIP analysis of ASH1L (296), MLL1 (456), H3K4me3, and H3K4me2 at (B) MLL1-independent genes and (C) MLL1-dependent genes in Mll1+/+ and _Mll1_−/− MEFs. Error bars represent standard deviations. 2.5 UR, 2.5 kb upstream region from the TSS; 0.5 and 2.0 TR, 0.5 and 2.0 kb downstream from the TSS, respectively; prom, promoter; rab Ig, rabbit immunoglobulin G.
Similar articles
- Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression.
Tanaka Y, Kawahashi K, Katagiri Z, Nakayama Y, Mahajan M, Kioussis D. Tanaka Y, et al. PLoS One. 2011;6(11):e28171. doi: 10.1371/journal.pone.0028171. Epub 2011 Nov 28. PLoS One. 2011. PMID: 22140534 Free PMC article. - ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia.
Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, Ikenouye L, Onishi M, Schneidawind D, Buechele C, Hansen L, Duque-Afonso J, Zhu F, Martin GM, Gozani O, Majeti R, Kutateladze TG, Cleary ML. Zhu L, et al. Cancer Discov. 2016 Jul;6(7):770-83. doi: 10.1158/2159-8290.CD-16-0058. Epub 2016 May 6. Cancer Discov. 2016. PMID: 27154821 Free PMC article. - Automethylation activities within the mixed lineage leukemia-1 (MLL1) core complex reveal evidence supporting a "two-active site" model for multiple histone H3 lysine 4 methylation.
Patel A, Vought VE, Swatkoski S, Viggiano S, Howard B, Dharmarajan V, Monteith KE, Kupakuwana G, Namitz KE, Shinsky SA, Cotter RJ, Cosgrove MS. Patel A, et al. J Biol Chem. 2014 Jan 10;289(2):868-84. doi: 10.1074/jbc.M113.501064. Epub 2013 Nov 14. J Biol Chem. 2014. PMID: 24235145 Free PMC article. - SET/MLL family proteins in hematopoiesis and leukemia.
Yang W, Ernst P. Yang W, et al. Int J Hematol. 2017 Jan;105(1):7-16. doi: 10.1007/s12185-016-2118-8. Epub 2016 Oct 31. Int J Hematol. 2017. PMID: 27796741 Review. - On your histone mark, SET, methylate!
Binda O. Binda O. Epigenetics. 2013 May;8(5):457-63. doi: 10.4161/epi.24451. Epub 2013 Apr 27. Epigenetics. 2013. PMID: 23625014 Free PMC article. Review.
Cited by
- Role of p53, Mitochondrial DNA Deletions, and Paternal Age in Autism: A Case-Control Study.
Wong S, Napoli E, Krakowiak P, Tassone F, Hertz-Picciotto I, Giulivi C. Wong S, et al. Pediatrics. 2016 Apr;137(4):e20151888. doi: 10.1542/peds.2015-1888. Epub 2016 Mar 31. Pediatrics. 2016. PMID: 27033107 Free PMC article. - Histone methyltransferases: regulation of transcription and contribution to human disease.
Nimura K, Ura K, Kaneda Y. Nimura K, et al. J Mol Med (Berl). 2010 Dec;88(12):1213-20. doi: 10.1007/s00109-010-0668-4. Epub 2010 Aug 17. J Mol Med (Berl). 2010. PMID: 20714703 Review. - Decreased SUV39H1 at the promoter region leads to increased CREMα and accelerates autoimmune response in CD4+ T cells from patients with systemic lupus erythematosus.
Luo S, Zhang H, Xie Y, Huang J, Luo D, Zhang Q. Luo S, et al. Clin Epigenetics. 2022 Dec 20;14(1):181. doi: 10.1186/s13148-022-01411-7. Clin Epigenetics. 2022. PMID: 36536372 Free PMC article. - Normal transcription of cellulolytic enzyme genes relies on the balance between the methylation of H3K36 and H3K4 in Penicillium oxalicum.
Li Y, Hu Y, Zhu Z, Zhao K, Liu G, Wang L, Qu Y, Zhao J, Qin Y. Li Y, et al. Biotechnol Biofuels. 2019 Aug 20;12:198. doi: 10.1186/s13068-019-1539-z. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31452679 Free PMC article. - Identification of Epigenetic Regulators of DUX4-fl for Targeted Therapy of Facioscapulohumeral Muscular Dystrophy.
Himeda CL, Jones TI, Virbasius CM, Zhu LJ, Green MR, Jones PL. Himeda CL, et al. Mol Ther. 2018 Jul 5;26(7):1797-1807. doi: 10.1016/j.ymthe.2018.04.019. Epub 2018 Apr 26. Mol Ther. 2018. PMID: 29759937 Free PMC article.
References
- Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280:17732-17736. - PubMed
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. - PubMed
- Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. - PubMed
- Cao, R., and Y. Zhang. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15:57-67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL007439/HL/NHLBI NIH HHS/United States
- 5P30CA016520-32/CA/NCI NIH HHS/United States
- R01 DK058044/DK/NIDDK NIH HHS/United States
- DK58044/DK/NIDDK NIH HHS/United States
- R37 DK058044/DK/NIDDK NIH HHS/United States
- R01 DK054937/DK/NIDDK NIH HHS/United States
- DK54937/DK/NIDDK NIH HHS/United States
- T32 HL07439-27/HL/NHLBI NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- T32 HL07439-26/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases