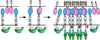Cell-cell signaling via Eph receptors and ephrins - PubMed (original) (raw)
Review
Cell-cell signaling via Eph receptors and ephrins
Juha-Pekka Himanen et al. Curr Opin Cell Biol. 2007 Oct.
Abstract
Eph receptors are the largest subfamily of receptor tyrosine kinases regulating cell shape, movements, and attachment. The interactions of the Ephs with their ephrin ligands are restricted to the sites of cell-cell contact since both molecules are membrane attached. This review summarizes recent advances in our understanding of the molecular mechanisms underlining the diverse functions of the molecules during development and in the adult organism. The unique properties of this signaling system that are of highest interest and have been the focus of intense investigations are as follows: (i) the signal is simultaneously transduced in both ligand-expressing cells and receptor-expressing cells, (ii) signaling via the same molecules can generate opposing cellular reactions depending on the context, and (iii) the Ephs and the ephrins are divided into two subclasses with promiscuous intrasubclass interactions, but rarely observed intersubclass interactions.
Figures
Figure 1
Activation of bi-directional Eph/ephrin signaling. The current model of signaling initiation involves an initial 1:1 high-affinity interaction between ligands and receptors, followed by tetramerization, oligomerization and clustering of the molecules at the sites of cell-cell contact [4]. This brings about phosphorylation of the cytoplasmic Eph (as well as B-class ephrin) domains and downstream signaling initiation [3]. Kinase activation is controlled by the phosphorylation of residues in the activation loop and the juxtamembrane segment, which affect the inter-lobe(subdomain) dynamics of the kinase domain [–35]. The minimal ligand-binding domain of the receptor is in blue and the ephrin receptor-binding domain is in pink. The kinase and SAM (sterile α-motif) domain is in green and the phosphate groups are in orange.
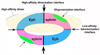
Figure 2
Schematic representation of the heterotetrameric complex formed between the Eph/ephrin interaction domains. Three interaction interfaces are indicated: dimerization and tetramerization - identified by crystallography [4] and confirmed by mutagenesis [25, 26], as well as a potential oligomerization interface - identified by a random mutagenesis approach [26].
Figure 3
Schematic representation and a comparison of the interaction surfaces in the canonical intra-subclass EphB2/ephrin-B2 [20] and EphB4/ephrin-B2 [24] complexes with the unusual inter-subclass EphB2/ephrin-A5 complex [21*]. While ephrin-B2 has several interacting surface areas with the B-class receptors, the interaction of ephrin-A5 with EphB2 is mostly confined to the hydrophobic receptor surface channel. Peptides “pep” [–41*] and small-molecule compounds “C” can also bind the channel, potentially inhibiting the Eph/ephrin interactions. The distinct architecture of the EphB2/ephrin-A5 complex and the lower affinity of the interaction can be used for the development of interaction assays, including FRET-based ones [22], suitable for high-throughput screening of compound libraries for small-molecule Eph signaling inhibitors “C”.
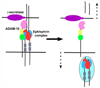
Figure 4
Cleavage of the Eph-bound ephrins allows for signaling termination and cell separation/repulsion. ADAM10 associates constitutively with the Eph receptors and ephrin binding generates a new ADAM-recognition motif at the Eph/ephrin interface allowing for productive protease positioning and efficient ephrin cleavage in trans. It seems likely that in certain cases cleavage in cis could also occur and that the Eph receptors could also potentially be cleaved off the cell surface. Further cleavage by a γ- secretase internalizes the cytoplasmic tails of the B-class ephrins initiating reverse signaling while the Eph/ephrin complexes are internalized in endosomes. The α- (ADAM10) and γ- secretases are likely retained at the cell surface to participate in another cleavage cycle. The interaction domains of the receptors are in blue and those of the ligands - in red. The protease domain of ADAM10 is in pink, the disintegrin domain - in yellow, and the Cys-rich domain - in green. The γ-secretase is drawn in purple.
Similar articles
- Eph receptor signaling and ephrins.
Lisabeth EM, Falivelli G, Pasquale EB. Lisabeth EM, et al. Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a009159. doi: 10.1101/cshperspect.a009159. Cold Spring Harb Perspect Biol. 2013. PMID: 24003208 Free PMC article. Review. - Eph signaling: a structural view.
Himanen JP, Nikolov DB. Himanen JP, et al. Trends Neurosci. 2003 Jan;26(1):46-51. doi: 10.1016/s0166-2236(02)00005-x. Trends Neurosci. 2003. PMID: 12495863 Review. - Eph receptors and ephrins.
Himanen JP, Nikolov DB. Himanen JP, et al. Int J Biochem Cell Biol. 2003 Feb;35(2):130-4. doi: 10.1016/s1357-2725(02)00096-1. Int J Biochem Cell Biol. 2003. PMID: 12479863 Review. - Mechanisms of boundary formation by Eph receptor and ephrin signaling.
Cayuso J, Xu Q, Wilkinson DG. Cayuso J, et al. Dev Biol. 2015 May 1;401(1):122-31. doi: 10.1016/j.ydbio.2014.11.013. Epub 2014 Nov 20. Dev Biol. 2015. PMID: 25448699 Review. - The Eph receptor/ephrin system: an emerging player in the invasion game.
Campbell TN, Robbins SM. Campbell TN, et al. Curr Issues Mol Biol. 2008;10(1-2):61-6. Curr Issues Mol Biol. 2008. PMID: 18525107 Review.
Cited by
- Ephrin-A1 and the sheddase ADAM12 are upregulated in COVID-19.
Mendoza R, Saha N, Momeni A, Gabutan E, Alawad M, Dehghani A, Diks J, Lin B, Wang D, Alshal M, Fyke W, Wang B, Himanen JP, Premsrirut P, Nikolov DB. Mendoza R, et al. Heliyon. 2021 Jun;7(6):e07200. doi: 10.1016/j.heliyon.2021.e07200. Epub 2021 May 31. Heliyon. 2021. PMID: 34095559 Free PMC article. - Human ribonuclease 1 serves as a secretory ligand of ephrin A4 receptor and induces breast tumor initiation.
Lee HH, Wang YN, Yang WH, Xia W, Wei Y, Chan LC, Wang YH, Jiang Z, Xu S, Yao J, Qiu Y, Hsu YH, Hwang WL, Yan M, Cha JH, Hsu JL, Shen J, Ye Y, Wu X, Hou MF, Tseng LM, Wang SC, Pan MR, Yang CH, Wang YL, Yamaguchi H, Pang D, Hortobagyi GN, Yu D, Hung MC. Lee HH, et al. Nat Commun. 2021 May 13;12(1):2788. doi: 10.1038/s41467-021-23075-2. Nat Commun. 2021. PMID: 33986289 Free PMC article. - Prognostic value of ephrin B receptors in breast cancer: An online survival analysis using the microarray data of 3,554 patients.
Mu X, Huang O, Jiang M, Xie Z, Chen D, Zhang X. Mu X, et al. Oncol Lett. 2019 Jul;18(1):742-750. doi: 10.3892/ol.2019.10363. Epub 2019 May 17. Oncol Lett. 2019. PMID: 31289549 Free PMC article. - Forward signaling by EphB1/EphB2 interacting with ephrin-B ligands at the optic chiasm is required to form the ipsilateral projection.
Chenaux G, Henkemeyer M. Chenaux G, et al. Eur J Neurosci. 2011 Nov;34(10):1620-33. doi: 10.1111/j.1460-9568.2011.07845.x. Eur J Neurosci. 2011. PMID: 22103419 Free PMC article. - The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors.
Tsuda H, Han SM, Yang Y, Tong C, Lin YQ, Mohan K, Haueter C, Zoghbi A, Harati Y, Kwan J, Miller MA, Bellen HJ. Tsuda H, et al. Cell. 2008 Jun 13;133(6):963-77. doi: 10.1016/j.cell.2008.04.039. Cell. 2008. PMID: 18555774 Free PMC article.
References
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005;6:462–475. - PubMed
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. - PubMed
- Kullander K, Klein R. Mechanism and function of Eph and ephrin signaling. Nature Rev.Mol.Cell Biol. 2002;3:475–486. - PubMed
- Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. - PubMed
- Holland SJ, et al. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722–725. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS038486-11A1/NS/NINDS NIH HHS/United States
- R01 NS038486/NS/NINDS NIH HHS/United States
- GM75886/GM/NIGMS NIH HHS/United States
- R21 GM075886/GM/NIGMS NIH HHS/United States
- NS38486/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous