Responses of hyperthermophilic crenarchaea to UV irradiation - PubMed (original) (raw)
Comparative Study
Responses of hyperthermophilic crenarchaea to UV irradiation
Dorothee Götz et al. Genome Biol. 2007.
Abstract
Background: DNA damage leads to cellular responses that include the increased expression of DNA repair genes, repression of DNA replication and alterations in cellular metabolism. Archaeal information processing pathways resemble those in eukaryotes, but archaeal damage response pathways remain poorly understood.
Results: We analyzed the transcriptional response to UV irradiation in two related crenarchaea, Sulfolobus solfataricus and Sulfolobus acidocaldarius. Sulfolobus species encounter high levels of DNA damage in nature, as they inhabit high temperature, aerobic environments and are exposed to sunlight. No increase in expression of DNA repair genes following UV irradiation was observed. There was, however, a clear transcriptional response, including repression of DNA replication and chromatin proteins. Differential effects on the expression of the three transcription factor B (tfb) genes hint at a mechanism for the modulation of transcriptional patterns in response to DNA damage. TFB3, which is strongly induced following UV irradiation, competes with TFB1 for binding to RNA polymerase in vitro, and may act as a repressor of transcription or an alternative transcription factor for certain promoters.
Conclusion: A clear response to DNA damage was observed, with down-regulation of the DNA replication machinery, changes in transcriptional regulatory proteins, and up-regulation of the biosynthetic enzymes for beta-carotene, which has UV protective properties, and proteins that detoxify reactive oxygen species. However, unlike eukaryotes and bacteria, there was no induction of DNA repair proteins in response to DNA damage, probably because these are expressed constitutively to deal with increased damage arising due to high growth temperatures.
Figures
Figure 1
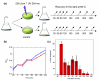
Experimental design and DNA damage. (a) Illustration of the experiment design. After growth to early log phase, cells were placed in Petri dishes and exposed to 200 J/m2 UV irradiation, then allowed to recover in the dark at 80°C. Arrows indicate sampling time points. (b) Optical density measurement of S. solfataricus UV-treated (red) and control (blue) cultures. Cells were exposed to UV at time zero. (c) Generation and removal of CPDs in chromosomal DNA. DNA damage was measured by ELISA using a primary antibody specific for CPDs. Data were normalized so that the signal at time zero (immediately after UV irradiation) represented 100% unrepaired CPDs. Each data point represents the mean from six experimental replicates and error bars show the standard deviation of each data set.
Figure 2
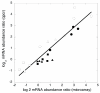
Comparison of transcript abundance quantified by microarray analysis and quantitative RT-PCR (qpcr). The changes in mRNA levels of six genes following UV irradiation were analyzed by quantitative RT-PCR to provide an independent confirmation of the microarray data. The log2 expression values for ssb (closed triangles), cdc6-2 (closed circles), tfb1 (closed squares), tfb2 (open squares), tfb3 (open circles) and dpoII (open triangles) obtained from microarray and RT-PCR analyses are plotted on the x and y axes, respectively. The data obtained from the two methods yield a linear fit with a slope of 0.93 and R-value of 0.91.
Figure 3
Cdc6-2 protein levels increase following UV irradiation. Western blot using antibodies against S. solfataricus Cdc6-1, Cdc6-2 and Cdc6-3. Cell-free extracts were prepared from samples taken from UV-treated and control cultures at the indicated time points. In agreement with the microarray data, the levels of Cdc6-2 protein show a clear increase after UV irradiation, whereas those of Cdc6-1 and Cdc6-3 are reduced.
Figure 4
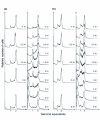
Flow cytometry DNA content distributions. Flow cytometry DNA content distributions for (a) S. solfataricus and (b) S. acidocaldarius from untreated control cultures (columns 1 and 3) and UV-irradiated cultures (columns 2 and 4) at different time points after treatment. The minor peak at a DNA content corresponding to 1.5 chromosome equivalents in the S. acidocaldarius distributions (columns 3 and 4) is a software artifact.
Figure 5
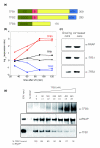
Differential effects of UV irradiation on expression of Sulfolobus tfb genes. (a) Both S. solfataricus and S. acidocaldarius encode three homologues of the basal transcription factor TFB. TFB1 and TFB2 are full-length orthologues, but TFB3 is severely truncated, lacking the B-finger domain (B) for transcription initiation and the HTH domain for binding to the BRE. C's represent the cysteines implicated in zinc binding (b) In both S. solfataricus and S. acidocaldarius, UV irradiation causes a dramatic increase in transcription of tfb3 and a significant reduction in expression of tfb2, whilst tfb1 levels are unaffected. Ratios of gene expression for the UV treated and control cultures are expressed in a log base 2 format, so that a value of 1 represents a two-fold increase in transcription. S. acidocaldarius and S. solfataricus data points are represented by triangles and circles, respectively. (c) Immunodetection of endogenous RNA polymerase (RNAP), TFB1 and TFB3 from cell lysates from control and UV-treated cells. Cells were harvested and lysed after 90 minutes of treatment. Levels of TFB3 protein increase after UV treatment in agreement with microarray data and quantitative RT-PCR (Table 2). (d) SDS-PAGE and western blot of immunoprecipitation of RNAP using specific antibodies (B' subunit) and co-immunoprecipitation of bound TFB1 and TFB3. Increasing concentrations of TFB3 (0-480 nM) were added to a mix with constant concentrations of RNAP and TFB1 (40 nM:80 nM). In the control lane where RNAP was not added, concentrations of TFB1 and TFB3 were 80 nM and 480 nM, respectively. The molar ratios of TFB3 and TFB1 are indicated below the TFB3 concentrations. Relative amounts of TFB1 bound to RNAP in each sample are indicated, showing a decrease of TFB1 bound to RNAP as TFB3 increases. All values are normalized with the total amount of RNAP immunoprecipitated in each sample.
Figure 6
The repair proteins RadA and SSB are not strongly induced by UV irradiation. Western blot using antibodies against S. solfataricus RadA and SSB. Cell-free extracts were prepared from samples taken from UV-treated and control cultures at the indicated time points. In agreement with the microarray data, RadA levels do not increase significantly following UV irradiation. The levels of SSB protein are also largely unchanged, in agreement with quantitative RT-PCR (Table 2), though the microarray data showed a modest rise in SSB transcription at early time points after UV damage.
Figure 7
Transcription of DNA repair genes is not induced significantly by UV irradiation. Known DNA repair genes are grouped by function, with time points for 30 minutes (white), 60 minutes (light grey), 90 minutes (dark grey) and 120 minutes (black) after irradiation shown. Ratios of gene expression for the UV-treated and control cultures are expressed in a log base 2 format, so that a value of 1 represents a two-fold increase in transcription and a value of -1 represents a two-fold decrease (indicated by horizontal grey lines). Few data points show an induction greater than two-fold. By contrast, lanes 22-24 show the highly induced genes cdc6-2, dpoII and tfb3, respectively. Repair genes are all from S. solfataricus (Table 1): 1, xpf; 2, xpb2; 3, fen1; 4, xpd; 5, radA; 6, hjc; 7, hje; 8, rad50; 9, mre11; 10, nurA; 11, herA; 12, ssb; 13, endoIII; 14, endoIV; 15, endoV; 16, mutT; 17, ogg1; 18, ogt; 19, udgaseIVa; 20, udgaseIVb; 21, udgaseVI. Data for S. acidocaldarius repair genes are qualitatively similar (Table 1).
Similar articles
- The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription.
Paytubi S, White MF. Paytubi S, et al. Mol Microbiol. 2009 Jun;72(6):1487-99. doi: 10.1111/j.1365-2958.2009.06737.x. Epub 2009 May 15. Mol Microbiol. 2009. PMID: 19460096 - Effect of UV irradiation on Sulfolobus acidocaldarius and involvement of the general transcription factor TFB3 in the early UV response.
Schult F, Le TN, Albersmeier A, Rauch B, Blumenkamp P, van der Does C, Goesmann A, Kalinowski J, Albers SV, Siebers B. Schult F, et al. Nucleic Acids Res. 2018 Aug 21;46(14):7179-7192. doi: 10.1093/nar/gky527. Nucleic Acids Res. 2018. PMID: 29982548 Free PMC article. - Participation of UV-regulated Genes in the Response to Helix-distorting DNA Damage in the Thermoacidophilic Crenarchaeon Sulfolobus acidocaldarius.
Suzuki S, Kurosawa N. Suzuki S, et al. Microbes Environ. 2019 Dec 27;34(4):363-373. doi: 10.1264/jsme2.ME19055. Epub 2019 Sep 21. Microbes Environ. 2019. PMID: 31548441 Free PMC article. - Reactions to UV damage in the model archaeon Sulfolobus solfataricus.
Fröls S, White MF, Schleper C. Fröls S, et al. Biochem Soc Trans. 2009 Feb;37(Pt 1):36-41. doi: 10.1042/BST0370036. Biochem Soc Trans. 2009. PMID: 19143598 Review. - Archaeal DNA repair: paradigms and puzzles.
White MF. White MF. Biochem Soc Trans. 2003 Jun;31(Pt 3):690-3. doi: 10.1042/bst0310690.. Biochem Soc Trans. 2003. PMID: 12773184 Review.
Cited by
- Driving Apart and Segregating Genomes in Archaea.
Barillà D. Barillà D. Trends Microbiol. 2016 Dec;24(12):957-967. doi: 10.1016/j.tim.2016.07.001. Epub 2016 Jul 20. Trends Microbiol. 2016. PMID: 27450111 Free PMC article. Review. - A Unique B-Family DNA Polymerase Facilitating Error-Prone DNA Damage Tolerance in Crenarchaeota.
Feng X, Liu X, Xu R, Zhao R, Feng W, Liao J, Han W, She Q. Feng X, et al. Front Microbiol. 2020 Jul 23;11:1585. doi: 10.3389/fmicb.2020.01585. eCollection 2020. Front Microbiol. 2020. PMID: 32793138 Free PMC article. - Hydroxyurea-Mediated Cytotoxicity Without Inhibition of Ribonucleotide Reductase.
Liew LP, Lim ZY, Cohen M, Kong Z, Marjavaara L, Chabes A, Bell SD. Liew LP, et al. Cell Rep. 2016 Nov 1;17(6):1657-1670. doi: 10.1016/j.celrep.2016.10.024. Cell Rep. 2016. PMID: 27806303 Free PMC article. - Replication-biased genome organisation in the crenarchaeon Sulfolobus.
Andersson AF, Pelve EA, Lindeberg S, Lundgren M, Nilsson P, Bernander R. Andersson AF, et al. BMC Genomics. 2010 Jul 28;11:454. doi: 10.1186/1471-2164-11-454. BMC Genomics. 2010. PMID: 20667100 Free PMC article. - Methionine Sulfoxide Reductases of Archaea.
Maupin-Furlow JA. Maupin-Furlow JA. Antioxidants (Basel). 2018 Sep 20;7(10):124. doi: 10.3390/antiox7100124. Antioxidants (Basel). 2018. PMID: 30241308 Free PMC article. Review.
References
- Kelman Z, White MF. Archaeal DNA replication and repair. Curr Opin Microbiol. 2005;8:669–676. - PubMed
- Grogan DW. Stability and repair of DNA in hyperthermophilic Archaea. Curr Issues Mol Biol. 2004;6:137–144. - PubMed
- Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In: Prakash L, Shermann F, Miller M, Lawrence C, Tabor HW, editor. Molecular and Environmental Aspects of Mutagenesis. Springfield, IL: Charles C Thomas; 1974.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources