Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I - PubMed (original) (raw)
Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I
Silke Hauf et al. EMBO J. 2007.
Abstract
Aurora-B kinases are important regulators of mitotic chromosome segregation, where they are required for the faithful bi-orientation of sister chromatids. In contrast to mitosis, sister chromatids have to be oriented toward the same spindle pole in meiosis-I, while homologous chromosomes are bi-oriented. We find that the fission yeast Aurora kinase Ark1 is required for the faithful bi-orientation of sister chromatids in mitosis and of homologous chromosomes in meiosis-I. Unexpectedly, Ark1 is also necessary for the faithful mono-orientation of sister chromatids in meiosis-I, even though the canonical mono-orientation pathway, which depends on Moa1 and Rec8, seems intact. Our data suggest that Ark1 prevents unified sister kinetochores during metaphase-I from merotelic attachment to both spindle poles and thus from being torn apart during anaphase-I, revealing a novel mechanism promoting monopolar attachment. Furthermore, our results provide an explanation for the previously enigmatic observation that fission yeast Shugoshin Sgo2, which assists in loading Aurora to centromeres, and its regulator Bub1 are required for the mono-orientation of sister chromatids in meiosis-I.
Figures
Figure 1
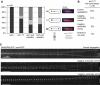
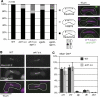
Ark1 is required for the bi-orientation of sister chromatids in mitosis. (A) Wild-type (ark1+), ark1-T7, ark1-as2 or ark1-as3 cells having _cen2_-GFP were synchronized in S-phase using hydroxyurea. Wild-type and ark1-T7 cells were shifted to the restrictive temperature for ark1-T7 (34°C) when being released, whereas ark1-as2 and ark1-as3 were grown at 30°C, but 5 μM 1NM-PP1 was added to the cultures at release. Eighty to hundred minutes after the release, the cells were fixed. Tubulin was visualized by TAT-1 antibody staining or mCherry-Atb2 (tubulin). cen2 segregation was determined in a minimum of 100 anaphase cells. (B, C) Ark1-T7 mutants having _cen2_-GFP and expressing Sid4-GFP to label the spindle pole bodies (SPB) were followed by live-cell microscopy at the restrictive temperature (34°C). Exemplary kymographs are shown in panel C.
Figure 2
Ark1 is required for the correction of malattachment. (A) The indicated strains carrying the nda3-KM311 mutation were arrested in mitosis by incubation at 19°C for 6 h, and released by transfer to 32°C. Where indicated (+inhib.) 5 μM 1NM-PP1 were added to the culture 10 min before release. Cells were fixed with methanol 10 min after upshift to 32°C. Chromosome segregation was assessed by determining _cen2_-GFP localization on anaphase spindles, which were labeled by mCherry-Atb2 (tubulin). (B, C) nda3-KM311 ark1-as3 cells were arrested in mitosis as in panel A and released by transferring cells to a microscope stage kept at 32°C. 1NM-PP1 (5 μM) was added 5 min before release. Only cells that could be followed through mitosis starting from very short spindle length were considered. Cells in which the spindle was defective and those in which centromere 2 did not attach to the spindle were excluded from the analysis. A kymograph of a cell that showed chromosome 2 segregating with one SPB is shown in panel C. Example kymographs for all phenotypes as well as example kymographs from ark1-as3 cells released from the nda3-KM311 arrest without any inhibitor are shown in Supplementary Figure S2A. (D) The indicated strains marked with _cen2_-GFP and mCherry-Atb2 (tubulin) were followed by live-cell microscopy at the restrictive temperature (34°C). A centromere was considered to have ‘switched' when the trajectory of _cen2_-GFP from one pole to the other could be clearly observed (arrows). The number of ‘switches' per hour in a total of 2 h prometaphase observation time is given.
Figure 3
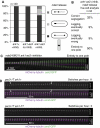
Ark1 is required for nuclear division during meiosis-I. (A) The position of metaphase-II spindles was evaluated in fixed cells. DNA was stained with Hoechst 33342. (B, C) The indicated strains were arrested after anaphase-I by the mes1 mutation. Cells were fixed and DNA was stained with DAPI. The number of nuclei was determined in at least 200 asci (C); exemplary cells are shown in panel B.
Figure 4
Ark1 is required for proper homolog bi-orientation in meiosis-I. (A) The indicated strains with both homologous chromosomes 2 marked by GFP were observed during anaphase-I using CFP-Atb2 (tubulin) as a marker. (B) The indicated strains, in which one of the homologous chromosomes 2 was marked by GFP, were fixed during meiosis by methanol. DNA was stained by Hoechst 33342 and the segregation of sister centromeres was determined in binucleated cells.
Figure 5
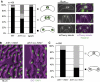
Ark1 is required for proper sister chromatid mono-orientation in meiosis-I independent of Moa1. (A) One of the homologous chromosomes 2 was marked by GFP in the indicated strains and the segregation pattern was determined during anaphase-I using CFP- or mCherry-Atb2 (tubulin) as a marker. (B) Cells from the indicated strains expressing Moa1-GFP and CFP-Atb2 (tubulin) were observed during metaphase-I. (C) Diploid strains of the indicated genotype were arrested in prophase-I by deletion of mei4+, and the amount of Moa1 at centromeres was determined by ChIP using a Moa1 antibody.
Figure 6
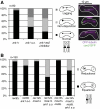
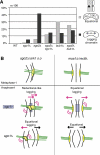
Sgo2 promotes mono-orientation similar to Ark1. (A) To assess the percentage of equational segregation (light gray bars), one of the homologous chromosomes 2 was marked by GFP in the indicated strains and the segregation pattern was judged in asci that had formed four spores. The percentage of lagging chromosomes or chromatids (dark gray bars) was determined by staining fixed cells with DAPI and the TAT-1 antibody recognizing tubulin. (B) Model for the chromosome segregation defects after Sgo2/Ark1 versus Moa1/Rec8 depletion. The centromeric region of one homolog is shown. In _sgo2_Δ or ark1 s.o. cells, the pair of sister kinetochores on one homolog tends to attach in a merotelic manner, that is, to both spindle poles, during metaphase-I. In anaphase, lagging homologs are common because of merotelic attachment. Eventually, both sister centromeres segregate to the same pole in most cases. If the protector Sgo1 is removed, sister centromeres segregate to opposite poles more frequently. Lagging chromatids are still observed after deletion of sgo1+, indicating merotelic attachment of single sister kinetochores. When _moa1_+ or _rec8_+ is deleted, sister kinetochores on one homolog lose their intimate connection. When _rec12_+ is deleted in addition to moa1+, sister chromatids segregate completely equationally in anaphase-I. Nevertheless, lagging chromosomes occur, but deleting _sgo1_+ abolishes their appearance.
Similar articles
- The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis.
Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. Kaitna S, et al. Curr Biol. 2002 May 14;12(10):798-812. doi: 10.1016/s0960-9822(02)00820-5. Curr Biol. 2002. PMID: 12015116 - Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I.
Sakuno T, Tanaka K, Hauf S, Watanabe Y. Sakuno T, et al. Dev Cell. 2011 Sep 13;21(3):534-45. doi: 10.1016/j.devcel.2011.08.012. Dev Cell. 2011. PMID: 21920317 - The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis.
Kitajima TS, Kawashima SA, Watanabe Y. Kitajima TS, et al. Nature. 2004 Feb 5;427(6974):510-7. doi: 10.1038/nature02312. Epub 2004 Jan 18. Nature. 2004. PMID: 14730319 - Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions.
Sakuno T, Watanabe Y. Sakuno T, et al. Chromosome Res. 2009;17(2):239-49. doi: 10.1007/s10577-008-9013-y. Chromosome Res. 2009. PMID: 19308704 Review. - Bi-orienting chromosomes on the mitotic spindle.
Tanaka TU. Tanaka TU. Curr Opin Cell Biol. 2002 Jun;14(3):365-71. doi: 10.1016/s0955-0674(02)00328-9. Curr Opin Cell Biol. 2002. PMID: 12067660 Review.
Cited by
- An interplay between Shugoshin and Spo13 for centromeric cohesin protection and sister kinetochore mono-orientation during meiosis I in Saccharomyces cerevisiae.
Mehta G, Anbalagan GK, Bharati AP, Gadre P, Ghosh SK. Mehta G, et al. Curr Genet. 2018 Oct;64(5):1141-1152. doi: 10.1007/s00294-018-0832-x. Epub 2018 Apr 11. Curr Genet. 2018. PMID: 29644457 - Chiasmata and the kinetochore component Dam1 are crucial for elimination of erroneous chromosome attachments and centromere oscillation at meiosis I.
Wakiya M, Nishi E, Kawai S, Yamada K, Katsumata K, Hirayasu A, Itabashi Y, Yamamoto A. Wakiya M, et al. Open Biol. 2021 Feb;11(2):200308. doi: 10.1098/rsob.200308. Epub 2021 Feb 3. Open Biol. 2021. PMID: 33529549 Free PMC article. - Meiosis in male Drosophila.
McKee BD, Yan R, Tsai JH. McKee BD, et al. Spermatogenesis. 2012 Jul 1;2(3):167-184. doi: 10.4161/spmg.21800. Spermatogenesis. 2012. PMID: 23087836 Free PMC article. - Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint.
Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. Shepperd LA, et al. Curr Biol. 2012 May 22;22(10):891-9. doi: 10.1016/j.cub.2012.03.051. Epub 2012 Apr 19. Curr Biol. 2012. PMID: 22521786 Free PMC article. - Different roles for Aurora B in condensin targeting during mitosis and meiosis.
Collette KS, Petty EL, Golenberg N, Bembenek JN, Csankovszki G. Collette KS, et al. J Cell Sci. 2011 Nov 1;124(Pt 21):3684-94. doi: 10.1242/jcs.088336. Epub 2011 Oct 24. J Cell Sci. 2011. PMID: 22025633 Free PMC article.
References
- Alfa C, Fantes P, Hyams J, McLeod M, Wabrick E (1993) Experiments with Fission Yeast. New York: Cold Spring Harbor Laboratory Press
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 - PubMed
- Bernard P, Maure JF, Javerzat JP (2001) Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol 3: 522–526 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous