V-ATPase V0 sector subunit a1 in neurons is a target of calmodulin - PubMed (original) (raw)
V-ATPase V0 sector subunit a1 in neurons is a target of calmodulin
Wei Zhang et al. J Biol Chem. 2008.
Abstract
The V(0) complex forms the proteolipid pore of a vesicular ATPase that acidifies vesicles. In addition, an independent function in membrane fusion has been suggested in vacuolar fusion in yeast and synaptic vesicle exocytosis in fly neurons. Evidence for a direct role in secretion has also recently been presented in mouse and worm. The molecular mechanisms of how the V(0) components might act or are regulated are largely unknown. Here we report the identification and characterization of a calmodulin-binding site in the large cytosolic N-terminal region of the Drosophila protein V100, the neuron-specific V(0) subunit a1. V100 forms a tight complex with calmodulin in a Ca(2+)-dependent manner. Mutations in the calmodulin-binding site in Drosophila lead to a loss of calmodulin recruitment to synapses. Neuronal expression of a calmodulin-binding deficient V100 uncovers an incomplete rescue at low levels and cellular toxicity at high levels. Our results suggest a vesicular ATPase V(0)-dependent function of calmodulin at synapses.
Figures
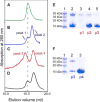
FIGURE 1. Characterization of binding of CaM to V100-N in a Ca2+ -dependent manner assessed by analytical gel filtration
Left row shows the following chromatograms: A, V100-N (0.06 mM); B, V100-N (0.06 mM) and CaM (0.15 mM) eluted with a buffer containing 2 mM EDTA; C, V100-N and CaM (same molar ratio as in B) with the mobile phase containing 5 mM Ca2+; D, identical to C, but with V100-N F319A mutant. In B, C, and D chromatograms, the molar ratio of V100-N to CaM is 1:2.5 with identical volume (0.25 ml) of each protein mixture loaded onto the column. Right row (E and F) shows the results of the SDS-PAGE and Coomassie Blue staining of the different fractions from the gel filtrations. The fractions were concentrated to roughly the same volume. p1–p4 correspond to the different elution peaks identified in the gel filtrations (B and C profiles). E, lane 1, protein markers; lane 2, a mixture of Ca2+ ·CaM and V100-N before loading; lane 3, peak 1; lane 4, first fraction from the peak of peak 2; lane 5, second fraction from the descending slope of peak 2. F, lane 1, protein markers; lane 2, peak 3; lane 3, peak 4.
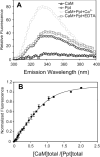
FIGURE 2. Binding of CaM to V100 CBD peptide assessed by Trp fluorescence
A, fluorescence emission spectra of CaM (0.3 _μ_M, triangle) and the CBD peptide (314NWFVKVRKIKAIYHTLNLFNLD338) of segment 2 (named Ppt) (0.3 _μ_M, diamond) and their complex in the presence of 5 mM Ca2+ (circle) or 10 mM EDTA (square). The spectra were obtained in 5 mM NaCl and 20 mM HEPES, pH 7.5 (see “Experimental Procedures”). The fluorescence emission was measured from 300 to 400 nm with the excitation wavelength set at 295 nm. B, fluorescence titration of Ca2+ ·CaM to V100 CBD peptide. The titration curve, using the normalized change in fluorescence at 340 nm, was fitted as described in “Experimental Procedures.” The fitting correlation coefficient (_R_2) is 0.98.
FIGURE 3. Alignment of the amino acid sequences of the CBD segments in the V-ATPase V0 subunit a in fly, human, mouse, worm, and yeast
The alignment is based on the alignment of the entire sequences of the various homologues. Residues in green background are invariant; yellow background, identical residues; cyan, similar residues. A, alignment of CBDs of only the a1 orthologs. B, alignment of the different subunit a orthologs. Sequence comparison of the fly subunit a homologues shows significantly high sequence identity among a1, a2, a4, and a5 (>50%), whereas a3 shares only 36% identity. Thus, there are only four major homologues (a1, a2, a4, and a5) in fly, which corresponds to the usual four orthologs present in other mammalian species. Worm Unc32, Vha5, Vha6, and Vha7 correspond to subunits a1, a2, a3, and a4, respectively. The yeast Vph1 segment is similar to those of subunit a1s largely due to the presence of the 1–10 and 1–14 pairs of hydrophobic residues, as well as Trp preceding the residue at position 1. However, the unusual presence of several acidic residues sets Vph1 apart from the other subunit a segments.
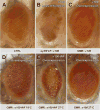
FIGURE 4. Expression of calmodulin-binding deficient V100 exhibits dosage-dependent cellular toxicity in Drosophila photoreceptors
A_–_C, pictures of a fly eye for control, loss, and gain of V100. All are morphologically normal. D_–_F, in contrast, overexpression of V100-WF causes mild developmental defects when expressed at low levels (by raising flies at 18 °C) and loss of photoreceptor neurons at high levels (27 °C). At the same high temperature, wild type v100 does not affect eye morphology (F). All effects were verified for three independent insertions of UAS-v100 and UAS-V100-WF.
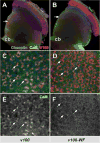
FIGURE 5. The CaM-binding domain in V100 is not required for V100 localization, but CaM is recruited to synapses
Shown are adult optic lobes from whole mount brain dissection showing photoreceptor projections in the brain. Photoreceptors are rendered mutant using the ey35FLP system and replaced with wild type (A, C, and E) or mutant (B, D, and F) V100. Blue, Chaoptin (24B10, a photoreceptor-specific marker); red, anti-V100; green, anti-CaM. A and B, in the optic lobes, V100 staining reveals the overexpressed V100 as well as V100-WF protein strongly enriched in synaptic layers (arrows), whereas Chaoptin (blue) primarily marks the axons. CaM is mostly detected in neuronal cell bodies (cb). C–F, high resolution lamina cross-section (upper arrows in A and B). V100 labels individual synaptic photoreceptor terminals. Only synaptic terminals enriched for wild type V100, but not V100-WF, exhibit increased levels of CaM at synapses (arrows).
Similar articles
- A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors.
Williamson WR, Wang D, Haberman AS, Hiesinger PR. Williamson WR, et al. J Cell Biol. 2010 May 31;189(5):885-99. doi: 10.1083/jcb.201003062. J Cell Biol. 2010. PMID: 20513768 Free PMC article. - Ca2+-Calmodulin regulates SNARE assembly and spontaneous neurotransmitter release via v-ATPase subunit V0a1.
Wang D, Epstein D, Khalaf O, Srinivasan S, Williamson WR, Fayyazuddin A, Quiocho FA, Hiesinger PR. Wang D, et al. J Cell Biol. 2014 Apr 14;205(1):21-31. doi: 10.1083/jcb.201312109. J Cell Biol. 2014. PMID: 24733584 Free PMC article. - The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila.
Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, Zhai RG, Verstreken P, Cao Y, Zhou Y, Kunz J, Bellen HJ. Hiesinger PR, et al. Cell. 2005 May 20;121(4):607-620. doi: 10.1016/j.cell.2005.03.012. Cell. 2005. PMID: 15907473 Free PMC article. - The membrane domain of vacuolar H(+)ATPase: a crucial player in neurotransmitter exocytotic release.
Morel N, Poëa-Guyon S. Morel N, et al. Cell Mol Life Sci. 2015 Jul;72(13):2561-73. doi: 10.1007/s00018-015-1886-2. Epub 2015 Mar 21. Cell Mol Life Sci. 2015. PMID: 25795337 Free PMC article. Review. - A new view of an old pore.
Bajjalieh S. Bajjalieh S. Cell. 2005 May 20;121(4):496-497. doi: 10.1016/j.cell.2005.05.002. Cell. 2005. PMID: 15907459 Review.
Cited by
- N-terminal domain of the V-ATPase a2-subunit displays integral membrane protein properties.
Merkulova M, McKee M, Dip PV, Grüber G, Marshansky V. Merkulova M, et al. Protein Sci. 2010 Oct;19(10):1850-62. doi: 10.1002/pro.470. Protein Sci. 2010. PMID: 20669186 Free PMC article. - Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice.
Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Dröse S, Brandt U, Savaskan E, Czech C, Götz J, Eckert A. Rhein V, et al. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):20057-62. doi: 10.1073/pnas.0905529106. Epub 2009 Nov 6. Proc Natl Acad Sci U S A. 2009. PMID: 19897719 Free PMC article. - Identification of domains within the V-ATPase accessory subunit Ac45 involved in V-ATPase transport and Ca2+-dependent exocytosis.
Jansen EJ, van Bakel NH, Olde Loohuis NF, Hafmans TG, Arentsen T, Coenen AJ, Scheenen WJ, Martens GJ. Jansen EJ, et al. J Biol Chem. 2012 Aug 10;287(33):27537-46. doi: 10.1074/jbc.M112.356105. Epub 2012 Jun 26. J Biol Chem. 2012. PMID: 22736765 Free PMC article. - The Human Mutation K237_V238del in a Putative Lipid Binding Motif within the V-ATPase a2 Isoform Suggests a Molecular Mechanism Underlying Cutis Laxa.
Chu A, Yao Y, Glibowicka M, Deber CM, Manolson MF. Chu A, et al. Int J Mol Sci. 2024 Feb 11;25(4):2170. doi: 10.3390/ijms25042170. Int J Mol Sci. 2024. PMID: 38396846 Free PMC article. - Clarifying lysosomal storage diseases.
Schultz ML, Tecedor L, Chang M, Davidson BL. Schultz ML, et al. Trends Neurosci. 2011 Aug;34(8):401-10. doi: 10.1016/j.tins.2011.05.006. Epub 2011 Jun 30. Trends Neurosci. 2011. PMID: 21723623 Free PMC article. Review.
References
- Chin D, Means AR. Trends Cell Biol. 2000;10:322–328. - PubMed
- Saimi Y, Kung C. Annu Rev Physiol. 2002;64:289–311. - PubMed
- Steinhardt RA, Alderton JM. Nature. 1982;295:154–155. - PubMed
- Nelson N, Harvey WR. Physiol Rev. 1999;79:361–385. - PubMed
- Nishi T, Forgac M. Nat Rev Mol Cell Biol. 2002;3:94–103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous