High-resolution structure prediction and the crystallographic phase problem - PubMed (original) (raw)
. 2007 Nov 8;450(7167):259-64.
doi: 10.1038/nature06249. Epub 2007 Oct 14.
Affiliations
- PMID: 17934447
- PMCID: PMC2504711
- DOI: 10.1038/nature06249
High-resolution structure prediction and the crystallographic phase problem
Bin Qian et al. Nature. 2007.
Abstract
The energy-based refinement of low-resolution protein structure models to atomic-level accuracy is a major challenge for computational structural biology. Here we describe a new approach to refining protein structure models that focuses sampling in regions most likely to contain errors while allowing the whole structure to relax in a physically realistic all-atom force field. In applications to models produced using nuclear magnetic resonance data and to comparative models based on distant structural homologues, the method can significantly improve the accuracy of the structures in terms of both the backbone conformations and the placement of core side chains. Furthermore, the resulting models satisfy a particularly stringent test: they provide significantly better solutions to the X-ray crystallographic phase problem in molecular replacement trials. Finally, we show that all-atom refinement can produce de novo protein structure predictions that reach the high accuracy required for molecular replacement without any experimental phase information and in the absence of templates suitable for molecular replacement from the Protein Data Bank. These results suggest that the combination of high-resolution structure prediction with state-of-the-art phasing tools may be unexpectedly powerful in phasing crystallographic data for which molecular replacement is hindered by the absence of sufficiently accurate previous models.
Figures
Figure 1
Overview of rebuilding-and-refinement method. a, Schematic diagram of the rebuilding-and-refinement method applied to structures from nuclear magnetic resonance (NMR), from comparative modeling (CM), and from de novo (DN) modeling approaches. b, Strong correlation between the per-residue backbone conformation variation in model ensemble and the deviation from the native structure for target T0199 from the Sixth Critical Assessment of Structure Prediction (CASP6). c, Superposition of the native structure of CASP6 target T0199 with fifty low energy all-atom refined models. The native structure backbone is shown as a thick line, and the models are shown as thinner lines. Residues of the native backbone structure are colored by the average per residue Cα root-mean-squared deviation (RMSD) to the native from 4.5Å (red) to 0.5 Å (blue). D. Iterative rebuilding and refinement yields low energy native-like models. The energy and the Cα-RMSD of models generated during three iterations of the loop-relax protocol are displayed for iteration 1 (green), iteration 4 (red), and iteration 7 (black). The Rosetta all-atom energy includes the enthalpy plus the solvation contribution to the entropy but not the configurational entropy.
Figure 2
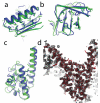
Improvement in model accuracy produced by rebuilding-and-refinement. a-d are NMR refinement tests, displaying superpositions of the crystal structure (blue), model 1 of NMR ensemble (red), and the all-atom refined model (green) for four NMR refinement test cases (a, acyl CoA binding protein 2abd; b, SH3 domain of ABL tyrosine kinase 1awo; c, guanine nucleotide binding protein 1ezy; and d, barstar 1ab7). e-h are blind predictions produced by comparative modeling, displaying superpositions of the native structure (blue), the best template in the Protein Data Bank (red), and the best of our five submitted models (green) for four CASP7 targets (e, T0380; f, T0385; g, T0330 domain 2; and h, T0331). A subset of the core side-chains are shown in stick representation to illustrate the accuracy of core packing. Figures were prepared in PyMOL (Delano Scientific, Palo Alto, CA).
Figure 3
Improvement in electron density using models from rebuilding-and-refinement in molecular replacement searches. Examples are presented for the NMR structure of acyl CoA binding protein 2abd (a and b) and CASP7 comparative modeling target T0385 (c and d). Black mesh represents electron density (2_m_Fo-_D_Fc; 1.5σ contour) using experimental structure factors and phases from molecular replacement with the starting model (a and c) or the refined model (b and d). The coordinates deposited in the Protein Data Bank, determined using experimental phase information, are shown in stick representation. Note that the “refinement” applied to the models refers to the all-atom energy-based protocol (see Figure 2 and text) and not to refinement against the diffraction data. The accurate modeling of side-chains by Rosetta was critical for the illustrated map improvement; molecular replacement trials gave significantly better solutions if the Rosetta-predicted side-chains were retained rather than truncated.
Figure 4
Ab initio phasing by ab initio modeling. a-c, Superpositions of blind Rosetta de novo structure predictions (green) and the subsequently released crystal structures (blue) for CASP7 targets T0354 (a), domain 3 of T0316 (b), and T0283 (c). Buried side-chains and backbone-aligned residues are displayed. d, Electron density map (2_m_Fo-_D_Fc; 2σ contour) produced by automatic refinement of the molecular replacement solution obtained from the T0283 structure prediction (black mesh; 1σ contour) agrees with the coordinates deposited in the Protein Data Bank (red), solved with experimental phase information. The electron density map immediately after molecular replacement is shown in Supplementary Figure S5.
Comment in
- Computational biology: protein predictions.
Dodson EJ. Dodson EJ. Nature. 2007 Nov 8;450(7167):176-7. doi: 10.1038/nature05990. Nature. 2007. PMID: 17934450 No abstract available.
Similar articles
- A database method for automated map interpretation in protein crystallography.
Diller DJ, Redinbo MR, Pohl E, Hol WG. Diller DJ, et al. Proteins. 1999 Sep 1;36(4):526-41. Proteins. 1999. PMID: 10450094 - Computational biology: protein predictions.
Dodson EJ. Dodson EJ. Nature. 2007 Nov 8;450(7167):176-7. doi: 10.1038/nature05990. Nature. 2007. PMID: 17934450 No abstract available. - Accelerating ab initio phasing with de novo models.
Shrestha R, Berenger F, Zhang KY. Shrestha R, et al. Acta Crystallogr D Biol Crystallogr. 2011 Sep;67(Pt 9):804-12. doi: 10.1107/S090744491102779X. Epub 2011 Aug 9. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21904033 - Implications of AlphaFold2 for crystallographic phasing by molecular replacement.
McCoy AJ, Sammito MD, Read RJ. McCoy AJ, et al. Acta Crystallogr D Struct Biol. 2022 Jan 1;78(Pt 1):1-13. doi: 10.1107/S2059798321012122. Epub 2022 Jan 1. Acta Crystallogr D Struct Biol. 2022. PMID: 34981757 Free PMC article. Review. - Acknowledging Errors: Advanced Molecular Replacement with Phaser.
McCoy AJ. McCoy AJ. Methods Mol Biol. 2017;1607:421-453. doi: 10.1007/978-1-4939-7000-1_18. Methods Mol Biol. 2017. PMID: 28573584 Review.
Cited by
- Efficient sampling of protein conformational space using fast loop building and batch minimization on highly parallel computers.
Tyka MD, Jung K, Baker D. Tyka MD, et al. J Comput Chem. 2012 Dec 5;33(31):2483-91. doi: 10.1002/jcc.23069. Epub 2012 Jul 27. J Comput Chem. 2012. PMID: 22847521 Free PMC article. - Targeted DNA methylation using an artificially bisected M.HhaI fused to zinc fingers.
Chaikind B, Kilambi KP, Gray JJ, Ostermeier M. Chaikind B, et al. PLoS One. 2012;7(9):e44852. doi: 10.1371/journal.pone.0044852. Epub 2012 Sep 11. PLoS One. 2012. PMID: 22984575 Free PMC article. - Challenges and surprises that arise with nucleic acids during model building and refinement.
Scott WG. Scott WG. Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):441-5. doi: 10.1107/S0907444912001084. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505264 Free PMC article. - Improved crystallographic models through iterated local density-guided model deformation and reciprocal-space refinement.
Terwilliger TC, Read RJ, Adams PD, Brunger AT, Afonine PV, Grosse-Kunstleve RW, Hung LW. Terwilliger TC, et al. Acta Crystallogr D Biol Crystallogr. 2012 Jul;68(Pt 7):861-70. doi: 10.1107/S0907444912015636. Epub 2012 Jun 19. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22751672 Free PMC article. - RosettaEPR: rotamer library for spin label structure and dynamics.
Alexander NS, Stein RA, Koteiche HA, Kaufmann KW, McHaourab HS, Meiler J. Alexander NS, et al. PLoS One. 2013 Sep 5;8(9):e72851. doi: 10.1371/journal.pone.0072851. eCollection 2013. PLoS One. 2013. PMID: 24039810 Free PMC article.
References
- Misura KM, Baker D. Progress and challenges in high-resolution refinement of protein structure models. Proteins. 2005;59:15–29. - PubMed
- Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–9. - PubMed
- Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2004;60:1229–36. - PubMed
- Giorgetti A, Raimondo D, Miele AE, Tramontano A. Evaluating the usefulness of protein structure models for molecular replacement. Bioinformatics. 2005;21(Suppl 2):ii72–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources