Essential role for Dicer during skeletal muscle development - PubMed (original) (raw)
Essential role for Dicer during skeletal muscle development
Jason R O'Rourke et al. Dev Biol. 2007.
Abstract
microRNAs (miRNAs) regulate gene expression post-transcriptionally by targeting mRNAs for degradation or by inhibiting translation. Dicer is an RNase III endonuclease which processes miRNA precursors into functional 21-23 nucleotide RNAs that are subsequently incorporated into the RNA-induced silencing complex. miRNA-mediated gene regulation is important for organogenesis of a variety of tissues including limb, lung and skin. To gain insight into the roles of Dicer and miRNAs in mammalian skeletal muscle development, we eliminated Dicer activity specifically in the myogenic compartment during embryogenesis. Dicer activity is essential for normal muscle development during embryogenesis and Dicer muscle mutants have reduced muscle miRNAs, die perinatally and display decreased skeletal muscle mass accompanied by abnormal myofiber morphology. Dicer mutant muscles also show increased apoptosis and Cre-mediated loss of Dicer in Myod-converted myoblasts results in enhanced cell death. These observations demonstrate key roles for Dicer in skeletal muscle and implicate miRNAs as critical components required for embryonic myogenesis.
Figures
Fig. 1
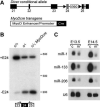
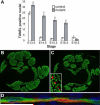
Inactivation of skeletal muscle Dicer and decreased muscle-specific miRNAs. (A) Dicer conditional allele and MyoDcre transgene. Dicer conditional allele contains loxP sites (triangles) flanking the RNAse III domain encoded by exon (filled boxes) 24 (gray box). A neomycin resistance cassette (neo) is retained in intron 24. MyoDcre transgene contains Cre recombinase (filled box) downstream of the MyoD core enhancer and proximal promoter elements (open box). (B) RT-PCR on E17.5 tongue cDNA using primers flanking targeted exon 24 (filled box). Primers (arrows) were positioned in exon 23 and 25. Mice contained a combination of Dicer conditional (c), wild type (+) or Dicer deleted exon 24 (−) alleles and a MyoDcre transgene. Transcripts containing (+E24) or missing exon 24 (−E24) result in 431 and 199 bp fragments, respectively. (C) RNA blot analysis of E13.5 and E14.5 control and mutant littermate limbs to detect muscle-specific miRNAs (miR). U6 snRNA is the loading control. Direct comparison between miR levels cannot be made because probe specific activities and film exposure times varied for each miR.
Fig. 2
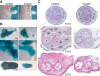
Dicer mutants display less skeletal muscle. (A) Skinned E18.5 control and mutant littermates. Note the impaired development of mutant forelimb muscle. (B) X-gal stained P0 control and mutant littermate forelimbs (top panels), hindlimbs (middle panels) and E18 tongues (bottom panels). Mice contained the R26 reporter in which X-gal specifically stains cells that express MyoDcre. (C) H&E staining of control and mutant transverse distal forelimb sections at different developmental time points (E13.5-E18.5). Each muscle is labeled in the E15.5 control section. Muscle and bones are brachioradials (BR), extensor carpi radialis brevis (ECR), extensor digitorum commusis (EDC), extensor digitorum lateralis (EDL), flexor carpi radialis (FCR), flexor carpi ulnaris (FCU), flexor digitorum profundus caput fadiale (FDP), pollicis longus (PL), and pronator teres (PT) muscles and ulna (U) and radius (R) bones.
Fig.3
Dicer mutants show skeletal muscle hypoplasia. (A) Desmin immunohistochemistry of control and mutant transverse distal forelimb sections at different developmental time points (E13.5-E18.5). Scale bars equal 250 μm. (B-C) Quantification of myofiber number (B) and cross sectional (C) area for each muscle group of E16.5 control and mutant littermate distal forelimbs. Asterisk signifies P<0.05. Muscle abbreviations are listed in Fig. 2 legend.
Fig. 4
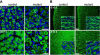
Abnormal myofiber morphology and organization in Dicer mutants. (A-B) Control and mutant littermate extensor carpi radialis transverse sections (A) or tongue sagittal sections (B) stained with antibody specific for desmin at E15.5 and E17.5. DAPI (blue) stains nuclei. Insets show representative higher magnification images of tongues. All scale bars equal 25 μm.
Fig. 5
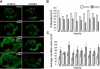
Increased apoptosis in Dicer mutant skeletal muscle. (A) Quantification of TUNEL positive nuclei in control and mutant littermate distal forelimb muscle masses at different developmental stages. At least three sections from each of two control and mutant embryos were counted for each time point. Please refer to Supplemental Fig. S2 for images. Asterisk indicates P<0.05. (B-C) Cleaved caspase-3 (red) and myosin heavy chain (green) coimmunohistochemistry on E15.5 control (B) and mutant littermate (C) transverse distal forelimb sections (10X). Inset (40X) shows higher magnification of a mutant section. (D) Cleaved caspase-3 (red) and myosin heavy chain (green) coimmunohistochemistry on E16.5 mutant sagittal distal forelimb section. Nuclei are stained with DAPI (blue).
Fig. 6
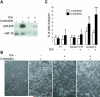
Decreased muscle miRNAs and increased apoptosis of Dicer mutant muscle cells in vitro. (A) RNA blot analysis of Dicer condtional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) fibroblast (− β-estradiol) and muscle cell (+ β-estradiol, 24 hrs.) cultures. miR-206 (which is induced by Myod-ER expression) and miR-16 (which is constitutively expressed in these cells and can be used as a loading control for first two lanes) were analyzed by stripping and sequential hybridization of the same blot. (B) Phase-contrast images of unfixed Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells, following 96 hours in DM with or without β-estradiol, as indicated. 10X magnification. (C) Flow cytometric analysis of Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells. Cells were cultured in DM with (+) or without β-estradiol (−) for 24 hours, as indicated. Cells were trypsinized, stained with Annexin V-FITC and propidium and sorted and quantified by FACS. Shown are summary data of the percentage of positively-stained cells, representing three independent trials. *: P<0.05, **: P<0.01 and ***: P<0.001
Similar articles
- Dicer activity in neural crest cells is essential for craniofacial organogenesis and pharyngeal arch artery morphogenesis.
Nie X, Wang Q, Jiao K. Nie X, et al. Mech Dev. 2011 Mar-Apr;128(3-4):200-7. doi: 10.1016/j.mod.2010.12.002. Epub 2011 Jan 21. Mech Dev. 2011. PMID: 21256960 Free PMC article. - miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation.
Saxena A, Tabin CJ. Saxena A, et al. Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):87-91. doi: 10.1073/pnas.0912870107. Epub 2009 Dec 14. Proc Natl Acad Sci U S A. 2010. PMID: 20018673 Free PMC article. - Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration.
Oikawa S, Lee M, Akimoto T. Oikawa S, et al. Int J Mol Sci. 2019 Nov 13;20(22):5686. doi: 10.3390/ijms20225686. Int J Mol Sci. 2019. PMID: 31766249 Free PMC article. - [Advance on Dicer gene and its role in female reproduction].
Li P, Zhu WJ. Li P, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011 Jun;28(3):275-8. doi: 10.3760/cma.j.issn.1003-9406.2011.03.008. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011. PMID: 21644222 Review. Chinese. - Piecing together the mosaic of early mammalian development through microRNAs.
Blakaj A, Lin H. Blakaj A, et al. J Biol Chem. 2008 Apr 11;283(15):9505-8. doi: 10.1074/jbc.R800002200. Epub 2008 Feb 13. J Biol Chem. 2008. PMID: 18272516 Free PMC article. Review.
Cited by
- Novel DICER1 mutation as cause of multinodular goiter in children.
Darrat I, Bedoyan JK, Chen M, Schuette JL, Lesperance MM. Darrat I, et al. Head Neck. 2013 Dec;35(12):E369-71. doi: 10.1002/hed.23250. Epub 2013 Jun 1. Head Neck. 2013. PMID: 23728841 Free PMC article. - RNA surveillance-an emerging role for RNA regulatory networks in aging.
Montano M, Long K. Montano M, et al. Ageing Res Rev. 2011 Apr;10(2):216-24. doi: 10.1016/j.arr.2010.02.002. Epub 2010 Feb 17. Ageing Res Rev. 2011. PMID: 20170753 Free PMC article. Review. - The Therapeutic Potential of Exosomes in Soft Tissue Repair and Regeneration.
Wan R, Hussain A, Behfar A, Moran SL, Zhao C. Wan R, et al. Int J Mol Sci. 2022 Mar 31;23(7):3869. doi: 10.3390/ijms23073869. Int J Mol Sci. 2022. PMID: 35409228 Free PMC article. Review. - Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway.
Wang Y, Ma J, Qiu W, Zhang J, Feng S, Zhou X, Wang X, Jin L, Long K, Liu L, Xiao W, Tang Q, Zhu L, Jiang Y, Li X, Li M. Wang Y, et al. Int J Mol Sci. 2018 Sep 19;19(9):2837. doi: 10.3390/ijms19092837. Int J Mol Sci. 2018. PMID: 30235878 Free PMC article. - MicroRNA-664-5p promotes myoblast proliferation and inhibits myoblast differentiation by targeting serum response factor and Wnt1.
Cai R, Qimuge N, Ma M, Wang Y, Tang G, Zhang Q, Sun Y, Chen X, Yu T, Dong W, Yang G, Pang W. Cai R, et al. J Biol Chem. 2018 Dec 14;293(50):19177-19190. doi: 10.1074/jbc.RA118.003198. Epub 2018 Oct 15. J Biol Chem. 2018. PMID: 30323063 Free PMC article.
References
- Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed
- Benezra R, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
- Bergstrom DA, et al. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG20911/AG/NIA NIH HHS/United States
- AR052581/AR/NIAMS NIH HHS/United States
- F32 AR052581/AR/NIAMS NIH HHS/United States
- F32 AR052581-02/AR/NIAMS NIH HHS/United States
- R01 AR046799/AR/NIAMS NIH HHS/United States
- R01 AG020911-05/AG/NIA NIH HHS/United States
- R01 AR046799-08/AR/NIAMS NIH HHS/United States
- R01 AR046799-07/AR/NIAMS NIH HHS/United States
- R01 AG020911/AG/NIA NIH HHS/United States
- AR46799/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases