Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components - PubMed (original) (raw)
Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components
Mahmoud El-Shami et al. Genes Dev. 2007.
Abstract
Two forms of RNA Polymerase IV (PolIVa/PolIVb) have been implicated in RNA-directed DNA methylation (RdDM) in Arabidopsis. Prevailing models imply a distinct function for PolIVb by association of Argonaute4 (AGO4) with the C-terminal domain (CTD) of its largest subunit NRPD1b. Here we show that the extended CTD of NRPD1b-type proteins exhibits conserved Argonaute-binding capacity through a WG/GW-rich region that functionally distinguishes Pol IVb from Pol IVa, and that is essential for RdDM. Site-specific mutagenesis and domain-swapping experiments between AtNRPD1b and the human protein GW182 demonstrated that reiterated WG/GW motifs form evolutionarily and functionally conserved Argonaute-binding platforms in RNA interference (RNAi)-related components.
Figures
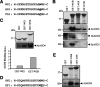
Figure 1.
The SD of the NRPD1b CTD binds AGO4 and is required for RdDM in Arabidopsis. (A) AtNRPD1a/b and NRPD1b/DSD proteins are shown schematically. The SD and CD are shown in red and green, respectively. The conserved box H and the CTD repeats are indicated. (B) Myc-AGO4 extract was applied onto equimolar amounts of GST and GST-based fusion protein beads, and the bound proteins were detected by immunoblotting with anti-Myc and anti-RPB1 antibodies. (C) AtSN1 DNA methylation in nrpd1b-11 is rescued by transgenes expressing NRPD1b but not NRPD1b/DSD (three T1 transgenic lines for each construct are shown). AtSN1 retroelement and controls (Cut 1 and control lanes) were amplified from HaeIII-digested genomic DNA as described (Pontier et al. 2005). (D) Proteins were extracted from pooled samples of the T1 transgenic lines shown in C, quantified, and subjected to immunoblot analysis using the anti-Flag M2 monoclonal antibodies (Sigma-Aldrich).
Figure 2.
The SD of the NRPD1b CTD has a WG/GW repeat signature sequence. (A) Dot plot analysis of AtNRPD1b versus SoNRPD1b highlighting the divergent region of the CTD. Positions of the divergent and AGO4-binding regions are indicated in relation to a schematic representation of AtNRPD1b. The SD shows sequence identity to the N-terminal region of HsGW182. The WG motifs are indicated with blue dots. The RNA recognition motif (RRM) domain of HsGW182 is indicated in brown. (B) Relative distribution of the WG/GW/GWG motifs within the SDs of plant NRPD1b proteins. (C) Myc-AGO4 or HA-AGO2 extracts were applied onto equimolar amounts of GST and GST-based fusion protein beads, and bound proteins were detected by immunoblotting with anti-Myc and anti-HA antibodies. (D) Alignment of AtNRPD1b and HsGW182. The WG/GW repeats are highlighted in red.
Figure 3.
Reiterated WG/GW motifs form conserved AGO-binding platform. (A) Sequences of wild-type and mutant AtNRPD1b consensus monomers. (B) Myc-AGO4 and HA-AGO1/2 extracts were applied onto equimolar amounts of GST and GST-based fusion protein beads, and bound proteins were detected by immunoblotting with anti-Myc and anti-HA antibodies. (C) Myc-AGO4 extract was applied onto equimolar amounts of GST, GST-WG1/WG8 beads, and bound proteins were detected by immunoblotting and quantified using ImageJ software. (D) Sequences of wild-type and mutant SoNRPD1b consensus monomers. (E) Myc-AGO4 extract was applied onto equimolar amounts of GST, GST-GW8/GF8 beads, and bound proteins were detected by immunoblotting with anti-Myc antibodies.
Figure 4.
Reiterated WG/GW motifs form functionally conserved AGO-binding platforms. (A) AtNRPD1b and NRPD1b/GW182 mutant are shown schematically. The N-terminal WG/GW-rich region of HsGW182 is shown in brown. (B) AtSN1 DNA methylation in nrpd1b-11 mutant is partially restored by transgenes expressing NRPD1b/GW182 (two T1 transgenic lines are shown). (C) 5S DNA methylation in nrpd1b-11 mutant is partially restored by transgenes expressing NRPD1b/GW182 (DNA was extracted from pooled samples of four T1 transgenic lines).
Figure 5.
Exhaustive search for conserved WG/GW platforms. Proteins containing conserved WG/GW motifs from different organisms were identified by PSI-BLAST alignment of the SD from AtNRPD1b against protein databases. An all-against-all alignment of the identified proteins was carried out using the Dotter program. The figure presents a twofold mirror image of the comparison of each sequence against all of the others, the diagonal representing self-versus-self alignments highlighting intramolecular repetitions. Sequences used were Arabidopsis thaliana NRPD1b; conceptual translations of Arabidopsis genes At5g04290 and At1g65440; P. tetraurelia; NOWA1/2; Homo sapiens proteins GW182, TNRC6B, and TNRC6C; Drosophila melanogaster protein GAWKY; T. thermophila cnjB; A. thaliana SDE3; C. elegans AIN-1; H. sapiens PrP27-30; and C. elegans PrP-like.
Similar articles
- NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein.
He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. He XJ, et al. Cell. 2009 May 1;137(3):498-508. doi: 10.1016/j.cell.2009.04.028. Cell. 2009. PMID: 19410546 Free PMC article. - PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit.
Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. Lahmy S, et al. Proc Natl Acad Sci U S A. 2009 Jan 20;106(3):941-6. doi: 10.1073/pnas.0810310106. Epub 2009 Jan 13. Proc Natl Acad Sci U S A. 2009. PMID: 19141635 Free PMC article. - Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing.
Haag JR, Pikaard CS. Haag JR, et al. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):483-92. doi: 10.1038/nrm3152. Nat Rev Mol Cell Biol. 2011. PMID: 21779025 Review. - RNA-directed DNA methylation and Pol IVb in Arabidopsis.
Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Matzke M, et al. Cold Spring Harb Symp Quant Biol. 2006;71:449-59. doi: 10.1101/sqb.2006.71.028. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381327 Review.
Cited by
- Mathematical model of RNA-directed DNA methylation predicts tuning of negative feedback required for stable maintenance.
Dale R, Mosher R. Dale R, et al. Open Biol. 2024 Nov;14(11):240159. doi: 10.1098/rsob.240159. Epub 2024 Nov 13. Open Biol. 2024. PMID: 39532148 Free PMC article. - Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - Mutations in the WG and GW motifs of the three RNA silencing suppressors of grapevine fanleaf virus alter their systemic suppression ability and affect virus infectivity.
Choi J, Browning S, Schmitt-Keichinger C, Fuchs M. Choi J, et al. Front Microbiol. 2024 Aug 12;15:1451285. doi: 10.3389/fmicb.2024.1451285. eCollection 2024. Front Microbiol. 2024. PMID: 39188317 Free PMC article. - Charophytic Green Algae Encode Ancestral Polymerase IV/Polymerase V Subunits and a CLSY/DRD1 Homolog.
Chakraborty T, Trujillo JT, Kendall T, Mosher RA. Chakraborty T, et al. Genome Biol Evol. 2024 Jun 4;16(6):evae119. doi: 10.1093/gbe/evae119. Genome Biol Evol. 2024. PMID: 38874416 Free PMC article. - Citrus psorosis virus 24K protein inhibits the processing of miRNA precursors by interacting with components of the biogenesis machinery.
Marmisolle FE, Borniego MB, Cambiagno DA, Gonzalo L, García ML, Manavella PA, Hernández C, Reyes CA. Marmisolle FE, et al. Microbiol Spectr. 2024 Jul 2;12(7):e0351323. doi: 10.1128/spectrum.03513-23. Epub 2024 May 24. Microbiol Spectr. 2024. PMID: 38785434 Free PMC article.
References
- Altschul S.F., Thomas L.D., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Thomas L.D., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Zhang J., Zhang Z., Miller W., Lipmann D.J., Zhang Z., Miller W., Lipmann D.J., Miller W., Lipmann D.J., Lipmann D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. - PMC - PubMed
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Doerks T., Stark A., Bork P., Izaurralde E., Stark A., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. - PMC - PubMed
- Brodersen P., Voinnet O., Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases