PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells - PubMed (original) (raw)
Review
doi: 10.1038/msb4100182. Epub 2007 Oct 16.
Johan Malmstrom, Bertran Gerrits, David Campbell, Henry Lam, Alexander Schmidt, Oliver Rinner, Lukas N Mueller, Paul T Shannon, Patrick G Pedrioli, Christian Panse, Hoo-Keun Lee, Ralph Schlapbach, Ruedi Aebersold
Affiliations
- PMID: 17940529
- PMCID: PMC2063582
- DOI: 10.1038/msb4100182
Review
PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells
Bernd Bodenmiller et al. Mol Syst Biol. 2007.
Abstract
The ability to analyze and understand the mechanisms by which cells process information is a key question of systems biology research. Such mechanisms critically depend on reversible phosphorylation of cellular proteins, a process that is catalyzed by protein kinases and phosphatases. Here, we present PhosphoPep, a database containing more than 10 000 unique high-confidence phosphorylation sites mapping to nearly 3500 gene models and 4600 distinct phosphoproteins of the Drosophila melanogaster Kc167 cell line. This constitutes the most comprehensive phosphorylation map of any single source to date. To enhance the utility of PhosphoPep, we also provide an array of software tools that allow users to browse through phosphorylation sites on single proteins or pathways, to easily integrate the data with other, external data types such as protein-protein interactions and to search the database via spectral matching. Finally, all data can be readily exported, for example, for targeted proteomics approaches and the data thus generated can be again validated using PhosphoPep, supporting iterative cycles of experimentation and analysis that are typical for systems biology research.
Figures
Figure 1
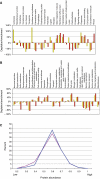
Phosphoprotein properties. (A) Depletion/enrichment of molecular functions derived from ‘panther' ontology (Mi et al, 2007) of the corresponding phosphoproteins (red) and the proteins identified from the separated peptides before enrichment (yellow) relative to the FlyBase database (0%) is shown. (B) Depletion/enrichment of biological functions derived from ‘panther' ontology (Mi et al, 2007) of the corresponding phosphoproteins (red) and the proteins identified from the separated peptides before enrichment (yellow) compared to the FlyBase database (0%) is shown. (C) A comparison of the predicted phosphoprotein abundance (blue) with the predicted abundance (Duret and Mouchiroud, 1999) of all proteins of the used FlyBase database (pink) is shown. The scale ranges from 0 (low abundance) to 1 (highly abundant). Proteins for which no molecular function or biological process could be assigned were omitted for (A) and (B). χ2 test results for (A) and (B) are shown in Supplementary Table II.
Figure 2
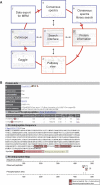
(A) Design of the PhosphoPep database. By using the ‘Search interface' (α) PhosphoPep can be interrogated for single proteins, a set of proteins or pathways. For each protein, several types of information including the observed phosphopeptides is shown in the ‘Protein information' page (see panel B and β. Single proteins or a set of proteins can be placed into their pathways (χ). From this ‘Pathway view' all phosphoproteins can be exported to Cytoscape (Shannon et al, 2003) (δ). This software tool allows integrating data from PhosphoPep with external data such as protein–protein interaction networks (ɛ). For most phosphopeptides, consensus MS2 spectra (φ) are given which can be exported for targeted proteomics experiments such as multiple reaction monitoring (Domon and Aebersold, 2006) (γ). As we supply an online spectral matching search tool, results generated by such experiments can be validated using PhosphoPep. (B) Representative output of the PhosphoPep database. The PhosphoPep (
) database contains more than 10 000 phosphorylation sites from nearly 3500 gene models and nearly 5800 phosphoproteins derived from the FlyBase (Grumbling and Strelets, 2006) nonredundant database (r4.3). For each phosphoprotein, the phosphopeptide sequence, the protein annotation and the predicted subcellular location is shown. Furthermore, additional information for each phosphopeptide is given: The probability, the number of tryptic ends, the dCn value, the mass, how often it was observed and to how many gene models and transcripts it maps. The phosphopeptides are represented in both the protein sequence and in a graphical representation, the protein map. Finally, a link to the ‘Pathway view', to the ‘Cytoscape export' function and to
(Obenauer et al, 2003) is given as represented by the three symbols besides the FlyBase gene entry.
Figure 3
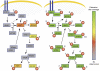
Proteins involved in the target of rapamycin (TOR) and insulin signaling. To demonstrate the usefulness of our database, we compared the already known phosphoproteins (left) with our identified phosphoproteins (right). As can be seen, compared to the literature in which only 6 out of the 15 proteins were found to be phosphorylated, we extended the phosphorylation map to all proteins of the pathway (Hay and Sonenberg, 2004; Oldham and Hafen, 2003) (phosphorylation sites are depicted by the P in a red circle, the number assigns the number of distinct phosphorylation sites). The number of identified phosphorylations ranged from 1 to 20 (CHICO). Peptides with _P_>0.8 and a defined phosphorylation site (dCn>0.1) were considered.
Similar articles
- Electron transfer dissociation in conjunction with collision activation to investigate the Drosophila melanogaster phosphoproteome.
Domon B, Bodenmiller B, Carapito C, Hao Z, Huehmer A, Aebersold R. Domon B, et al. J Proteome Res. 2009 Jun;8(6):2633-9. doi: 10.1021/pr800834e. J Proteome Res. 2009. PMID: 19435317 - Phosphoproteome resource for systems biology research.
Bodenmiller B, Aebersold R. Bodenmiller B, et al. Methods Mol Biol. 2011;694:307-22. doi: 10.1007/978-1-60761-977-2_19. Methods Mol Biol. 2011. PMID: 21082442 - An integrated chemical, mass spectrometric and computational strategy for (quantitative) phosphoproteomics: application to Drosophila melanogaster Kc167 cells.
Bodenmiller B, Mueller LN, Pedrioli PG, Pflieger D, Jünger MA, Eng JK, Aebersold R, Tao WA. Bodenmiller B, et al. Mol Biosyst. 2007 Apr;3(4):275-86. doi: 10.1039/b617545g. Epub 2007 Feb 19. Mol Biosyst. 2007. PMID: 17372656 - Mass spectrometry-driven phosphoproteomics: patterning the systems biology mosaic.
Jünger MA, Aebersold R. Jünger MA, et al. Wiley Interdiscip Rev Dev Biol. 2014 Jan-Feb;3(1):83-112. doi: 10.1002/wdev.121. Epub 2013 Jul 2. Wiley Interdiscip Rev Dev Biol. 2014. PMID: 24902836 Review. - Phosphoinositides at the neuromuscular junction of Drosophila melanogaster: a genetic approach.
Slabbaert JR, Khuong TM, Verstreken P. Slabbaert JR, et al. Methods Cell Biol. 2012;108:227-47. doi: 10.1016/B978-0-12-386487-1.00012-2. Methods Cell Biol. 2012. PMID: 22325606 Review.
Cited by
- Comprehensive Overview of Bottom-Up Proteomics Using Mass Spectrometry.
Jiang Y, Rex DAB, Schuster D, Neely BA, Rosano GL, Volkmar N, Momenzadeh A, Peters-Clarke TM, Egbert SB, Kreimer S, Doud EH, Crook OM, Yadav AK, Vanuopadath M, Hegeman AD, Mayta ML, Duboff AG, Riley NM, Moritz RL, Meyer JG. Jiang Y, et al. ACS Meas Sci Au. 2024 Jun 4;4(4):338-417. doi: 10.1021/acsmeasuresciau.3c00068. eCollection 2024 Aug 21. ACS Meas Sci Au. 2024. PMID: 39193565 Free PMC article. Review. - A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila.
Chen CC, Greene E, Bowers SR, Mellone BG. Chen CC, et al. PLoS One. 2012;7(9):e45094. doi: 10.1371/journal.pone.0045094. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23028777 Free PMC article. - mei-38 is required for chromosome segregation during meiosis in Drosophila females.
Wu C, Singaram V, McKim KS. Wu C, et al. Genetics. 2008 Sep;180(1):61-72. doi: 10.1534/genetics.108.091140. Epub 2008 Aug 30. Genetics. 2008. PMID: 18757915 Free PMC article. - Optimization of TripleTOF spectral simulation and library searching for confident localization of phosphorylation sites.
Takai A, Tsubosaka T, Hirano Y, Hayakawa N, Tani F, Haapaniemi P, Suni V, Imanishi SY. Takai A, et al. PLoS One. 2019 Dec 2;14(12):e0225885. doi: 10.1371/journal.pone.0225885. eCollection 2019. PLoS One. 2019. PMID: 31790495 Free PMC article. - Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control.
Jansen JM, Wanless AG, Seidel CW, Weiss EL. Jansen JM, et al. Curr Biol. 2009 Dec 29;19(24):2114-20. doi: 10.1016/j.cub.2009.10.071. Epub 2009 Dec 3. Curr Biol. 2009. PMID: 19962308 Free PMC article.
References
- Aebersold R, Goodlett DR (2001) Mass spectrometry in proteomics. Chem Rev 101: 269–295 - PubMed
- Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422: 198–207 - PubMed
- Andersson L, Porath J (1986) Isolation of phosphoproteins by immobilized metal (Fe3+) affinity-chromatography. Anal Biochem 154: 250–254 - PubMed
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24: 1285–1292 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases