Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity - PubMed (original) (raw)
Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity
Yueh-Ming Loo et al. J Virol. 2008 Jan.
Abstract
Alpha/beta interferon immune defenses are essential for resistance to viruses and can be triggered through the actions of the cytoplasmic helicases retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5). Signaling by each is initiated by the recognition of viral products such as RNA and occurs through downstream interaction with the IPS-1 adaptor protein. We directly compared the innate immune signaling requirements of representative viruses of the Flaviviridae, Orthomyxoviridae, Paramyxoviridae, and Reoviridae for RIG-I, MDA5, and interferon promoter-stimulating factor 1 (IPS-1). In cultured fibroblasts, IPS-1 was essential for innate immune signaling of downstream interferon regulatory factor 3 activation and interferon-stimulated gene expression, but the requirements for RIG-I and MDA5 were variable. Each was individually dispensable for signaling triggered by reovirus and dengue virus, whereas RIG-I was essential for signaling by influenza A virus, influenza B virus, and human respiratory syncytial virus. Functional genomics analyses identified cellular genes triggered during influenza A virus infection whose expression was strictly dependent on RIG-I and which are involved in processes of innate or adaptive immunity, apoptosis, cytokine signaling, and inflammation associated with the host response to contemporary and pandemic strains of influenza virus. These results define IPS-1-dependent signaling as an essential feature of host immunity to RNA virus infection. Our observations further demonstrate differential and redundant roles for RIG-I and MDA5 in pathogen recognition and innate immune signaling that may reflect unique and shared biologic properties of RNA viruses whose differential triggering and control of gene expression may impact pathogenesis and infection.
Figures
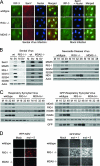
FIG. 1.
RIG-I-dependent signaling of the innate immune response during paramyxovirus infection. (A) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (right panels) or infected with SenV at 100 HA units/ml of medium (left panels) for 12 h. Cells were fixed and stained with primary antibodies specific for IRF-3 and SenV, followed by fluorescent dye-conjugated secondary antibodies. Cellular distribution of IRF-3 (green), SenV gene products (red), and 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (blue) was visualized by immunofluorescence microscopy. (B) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with SenV (SV) at 100 HA units/ml of medium for 24 h (left) or with NDV at a multiplicity of infection (MOI) of 5 for the indicated times (hours) (right). (C) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with RSV (left) or GFP-RSV (right) at an MOI of 5 for the indicated times (hours). Cells were collected and analyzed by immunoblotting for expression of ISG15, ISG54, ISG56, RIG-I, MDA5, viral proteins, and actin (used as a control). (D) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with either RFP-NDV (left) or GFP-RSV (right) at the indicated MOI for 48 h; then they were fixed, and virus replication was analyzed using fluorescence microscopy.
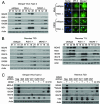
FIG. 2.
DEN and reoviruses trigger the innate immune response independently of RIG-I or MDA5. (A) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with DEN2 at a multiplicity of infection of 1. (Left) Extracts derived from cells that were mock infected (M) or infected with DEN2 for the indicated times (in hours) were analyzed by immunoblotting for the abundance of IRF-3-responsive genes, RIG-I, MDA5, DEN2 NS1, and actin (used as a control). (Right) At 16 h postinfection, cells were fixed and stained with primary antibodies specific for IRF-3, followed by fluorescent-dye-conjugated secondary antibodies. Cellular distribution of IRF-3 (green) and 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (blue) was analyzed by immunofluorescence microscopy. (B) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with either reovirus T3D or reovirus T1L at a multiplicity of infection of 25. At the indicated times postinfection (in hours), cells were harvested and the extracts analyzed by immunoblotting. We note that a significantly longer exposure time was necessary for the detection of ISG54 and ISG56 in reovirus T1L-infected cells. (C) RIG-I−/− mouse fibroblasts were infected with lentivirus particles expressing either a shRNA to MDA5 (shRNA 998, 2067, 2911, 3265, or 3547) or a nontargeting shRNA control (Neg). At 36 h postinfection, cells were either mock infected (−) or infected (+) with either DEN (left) or reovirus T3D (right) at multiplicities of infection of 1 and 25, respectively. Cells were harvested 48 h postinfection (84 h after infection with lentivirus) and the extracts analyzed by immunoblotting. For controls, extracts of RIG-I−/− cells that were mock treated (M) or treated with exogenous IFN-β were also subjected to immunoblot analysis. **MDA5 indicates an image of MDA5 protein expression taken after a significantly longer exposure time.
FIG. 3.
IPS-1 is essential for triggering of the innate immune response during paramyxovirus, reovirus, or DEN infection. (A) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected with SenV at 100 HA units/ml. (Left) Cells were fixed and stained with primary and fluorescent-dye-conjugated secondary antibodies at 12 h postinfection. The cellular distribution of IRF-3 (green), SenV-infected cells (red), and nuclei of cells (blue) (stained with 4′,6′-diamidino-2-phenylindole [DAPI]) was detected by fluorescence microscopy. (Right) At 24 h, mock-infected (M) and SenV-infected (SV) cells were collected and analyzed by immunoblotting for the abundances of ISG54, RIG-I, MDA5, IPS-1, viral antigen, and actin (used as a control). (B) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected (V) with either reovirus T3D at a multiplicity of infection of 25 or DEN2 at a multiplicity of infection of 1. At 24 h postinfection, cells were collected and analyzed for the abundances of RIG-I, MDA5, ISG54, viral antigen, and actin (used as a control). (C) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected with DEN2 at a multiplicity of infection of 1 for 24 h. Cells were fixed, stained with primary and fluorescent-dye-conjugated secondary antibodies, and analyzed by immunofluorescent microscopy for the cellular distribution of IRF-3 (green) and the detection of DEN2-infected cells (red) and DAPI-stained nuclei (blue). (D) HeLa cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids and either a siRNA pool that is known to suppress IPS-1 or RIG-I expression or a nonspecific control siRNA (scrambled). At 24 h posttransfection, cells were either mock infected or infected with either SenV at 100 HA units/ml or DEN2 at a multiplicity of infection of 1. Cells were collected at 24 h postinfection and analyzed for relative luciferase activity. Error bars, standard deviations calculated from three independent experiments.
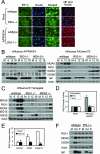
FIG. 4.
RIG-I- and IPS-1-dependent innate immune signaling by orthomyxoviruses. (A) (Left panels) wt or RIG-I−/− mouse fibroblasts were either mock infected or infected with influenza virus A/PR/8/34 at a multiplicity of infection of 5. At 12 h postinfection, cells were fixed and analyzed for host response activation by staining for IRF-3 (green) and nuclei (blue). (Right panel) In parallel experiments, cells were fixed and analyzed for the frequency of virus infection by staining for influenza A virus NP (red) and nuclei (blue). (B and C) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with influenza virus A/PR/8/34 or A/Udorn/72 (B) or with influenza virus B/Yamagata (C) at a multiplicity of infection of 5. At the indicated hours postinfection, cells were collected and analyzed by immunoblotting for the abundances of IRF-3-responsive genes, MDA5, RIG-I, influenza virus NP or NS1B protein, and actin (used as a control). (D) HeLa cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids and either a siRNA pool that has been shown to suppress IPS-1 or RIG-I expression or a control siRNA with no known targets (Scrambled). At 24 h posttransfection, cells were either mock infected or infected with influenza virus A/PR/8delNS1 at a multiplicity of infection of 5. Cells were collected 18 h postinfection and analyzed for dual luciferase activity(expressed as relative IFN-β promoter activity). Error bars, standard deviations calculated from three independent experiments. (E) Huh7 or Huh7.5 cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids for 24 h and then were either mock transfected or transfected with either vRNA or a synthetic double-stranded RNA control [poly(I:C)]. At 12 h after RNA transfection (36 h after plasmid DNA transfection), cells were collected and analyzed for dual luciferase activity. The results are expressed as relative IFN-β promoter activity. Error bars, standard deviations calculated from three independent experiments. (F) wt or IPS-1−/− mouse fibroblasts were either mock infected (M) or infected with influenza virus A/Udorn/72 (Ud) or A/PR/8/34 (P) at a multiplicity of infection of 5. At 18 h postinfection, cells were collected and the extracts analyzed for the abundances of RIG-I, MDA5, ISG54, actin (used as a control), and viral NP by immunoblotting.
FIG. 5.
Identification of RIG-I-responsive genes by functional genomics analyses. wt or RIG-I−/− mouse fibroblasts were either mock infected or infected with influenza virus A/PR/8/34 at a multiplicity of infection of 5. At 8 and 26 h postinfection, cells were collected and cellular RNA extracted for DNA microarray or qPCR analyses as described in the text. (A) (Top) Numbers of differentially expressed genes (up- or down-regulated more than twofold) as identified by DNA microarray analyses in wt and RIG-I−/− cells following influenza virus A/PR/8/34 infection at 8 and 26 h relative to expression in mock-infected cells. (Bottom) Table showing the total numbers of differentially expressed genes and the breakdown of genes that were differentially expressed as a percentage of total genes analyzed. (B) Heat map showing the differential expression of a bioset of RIG-I-responsive genes following influenza virus A/PR/8/34 infection. (C) Gene expression of IFN-α6 and IFN-β was verified by qualitative real-time PCR. (D) Numbers of genes, segregated by their known functions, whose expression was determined by a DNA microarray to be induced more than twofold following influenza virus A/PR/8/34 infection for 8 and 26 h. Additional data are available at
http://expression.viromics.washington.edu
. (E) Qualitative real-time PCR analysis for viral HA protein and NP in RNA extracted from wt or RIG-I−/− cells that were either mock infected or infected with influenza virus A/PR/8/34 for 8 or 26 h.
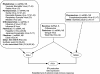
FIG. 6.
Model of viral nucleic acid pathogen-associated molecular pattern recognition and signaling to IFN-β by RIG-I and MDA5 during infection by different RNA viruses. Shown is a summary of published results (with reference numbers in parentheses) demonstrating virus-induced RIG-I and MDA5 signaling of the innate immune response. Genomic features of viruses are described in brief: positive sense (+) versus negative sense (−); single-stranded RNA (ssRNA) versus double-stranded RNA (dsRNA); segmented (S) versus nonsegmented (NS) genomes. In fibroblasts, epithelial cells, and certain immune cells, signaling by RIG-I and MDA5 converges on IPS-1 to rapidly trigger IFN-α/β production and ISG expression. Of the viruses listed, reovirus, DEN2, and West Nile virus are unique in being able to signal both RIG-I-dependent and RIG-I-independent responses.
Similar articles
- Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1.
Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M Jr. Fredericksen BL, et al. J Virol. 2008 Jan;82(2):609-16. doi: 10.1128/JVI.01305-07. Epub 2007 Oct 31. J Virol. 2008. PMID: 17977974 Free PMC article. - Essential role of IPS-1 in innate immune responses against RNA viruses.
Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Kumar H, et al. J Exp Med. 2006 Jul 10;203(7):1795-803. doi: 10.1084/jem.20060792. Epub 2006 Jun 19. J Exp Med. 2006. PMID: 16785313 Free PMC article. - Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Kato H, et al. Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202 - Regulation of RLR-mediated innate immune signaling--it is all about keeping the balance.
Eisenächer K, Krug A. Eisenächer K, et al. Eur J Cell Biol. 2012 Jan;91(1):36-47. doi: 10.1016/j.ejcb.2011.01.011. Epub 2011 Apr 9. Eur J Cell Biol. 2012. PMID: 21481967 Review. - RIG-I in RNA virus recognition.
Kell AM, Gale M Jr. Kell AM, et al. Virology. 2015 May;479-480:110-21. doi: 10.1016/j.virol.2015.02.017. Epub 2015 Mar 5. Virology. 2015. PMID: 25749629 Free PMC article. Review.
Cited by
- An attenuating mutation in a neurovirulent Sindbis virus strain interacts with the IPS-1 signaling pathway in vivo.
Wollish AC, Ferris MT, Blevins LK, Loo YM, Gale M Jr, Heise MT. Wollish AC, et al. Virology. 2013 Jan 20;435(2):269-80. doi: 10.1016/j.virol.2012.09.008. Epub 2012 Oct 17. Virology. 2013. PMID: 23084425 Free PMC article. - The Atypical Kinase RIOK3 Limits RVFV Propagation and Is Regulated by Alternative Splicing.
Havranek KE, White LA, Bisom TC, Lanchy JM, Lodmell JS. Havranek KE, et al. Viruses. 2021 Feb 26;13(3):367. doi: 10.3390/v13030367. Viruses. 2021. PMID: 33652597 Free PMC article. - Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction.
Maharaj NP, Wies E, Stoll A, Gack MU. Maharaj NP, et al. J Virol. 2012 Feb;86(3):1358-71. doi: 10.1128/JVI.06543-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114345 Free PMC article. - The Impacts of Antivirals on the Coronavirus Genome Structure and Subsequent Pathogenicity, Virus Fitness and Antiviral Design.
Lin CH, Yang CY, Ou SC, Wang M, Lo CY, Tsai TL, Wu HY. Lin CH, et al. Biomedicines. 2020 Sep 24;8(10):376. doi: 10.3390/biomedicines8100376. Biomedicines. 2020. PMID: 32987828 Free PMC article. - DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication.
Fullam A, Schröder M. Fullam A, et al. Biochim Biophys Acta. 2013 Aug;1829(8):854-65. doi: 10.1016/j.bbagrm.2013.03.012. Epub 2013 Apr 6. Biochim Biophys Acta. 2013. PMID: 23567047 Free PMC article. Review.
References
- Barthold, S. W., A. L. Smith, and P. N. Bhatt. 1993. Infectivity, disease patterns, and serologic profiles of reovirus serotypes 1, 2, and 3 in infant and weanling mice. Lab. Anim. Sci. 43425-430. - PubMed
- Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29365-371. - PubMed
- CDC. 2006. Respiratory syncytial virus activity—United States, 2005-2006. Morb. Mortal. Wkly. Rep. 551277-1279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI060389/AI/NIAID NIH HHS/United States
- R01 AI22646/AI/NIAID NIH HHS/United States
- U19 AI62623/AI/NIAID NIH HHS/United States
- R01 AI022646/AI/NIAID NIH HHS/United States
- CA09229-28/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U19 AI062623/AI/NIAID NIH HHS/United States
- R01 AI46954/AI/NIAID NIH HHS/United States
- R01 AI060389/AI/NIAID NIH HHS/United States
- T32 CA009229/CA/NCI NIH HHS/United States
- R01 AI046954/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous