Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses - PubMed (original) (raw)
Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses
Gisle Vestergaard et al. J Virol. 2008 Jan.
Abstract
Four novel filamentous viruses with double-stranded DNA genomes, namely, Acidianus filamentous virus 3 (AFV3), AFV6, AFV7, and AFV8, have been characterized from the hyperthermophilic archaeal genus Acidianus, and they are assigned to the Betalipothrixvirus genus of the family Lipothrixviridae. The structures of the approximately 2-mum-long virions are similar, and one of them, AFV3, was studied in detail. It consists of a cylindrical envelope containing globular subunits arranged in a helical formation that is unique for any known double-stranded DNA virus. The envelope is 3.1 nm thick and encases an inner core with two parallel rows of protein subunits arranged like a zipper. Each end of the virion is tapered and carries three short filaments. Two major structural proteins were identified as being common to all betalipothrixviruses. The viral genomes were sequenced and analyzed, and they reveal a high level of conservation in both gene content and gene order over large regions, with this similarity extending partly to the earlier described betalipothrixvirus Sulfolobus islandicus filamentous virus. A few predicted gene products of each virus, in addition to the structural proteins, could be assigned specific functions, including a putative helicase involved in Holliday junction branch migration, a nuclease, a protein phosphatase, transcriptional regulators, and glycosyltransferases. The AFV7 genome appears to have undergone intergenomic recombination with a large section of an AFV2-like viral genome, apparently resulting in phenotypic changes, as revealed by the presence of AFV2-like termini in the AFV7 virions. Shared features of the genomes include (i) large inverted terminal repeats exhibiting conserved, regularly spaced direct repeats; (ii) a highly conserved operon encoding the two major structural proteins; (iii) multiple overlapping open reading frames, which may be indicative of gene recoding; (iv) putative 12-bp genetic elements; and (v) partial gene sequences corresponding closely to spacer sequences of chromosomal repeat clusters.
Figures
FIG. 1.
Electron micrographs of 2-μm-long filamentous viruses from a hot spring at Pozzuoli, Italy. (A) AFV3 replicated in Acidianus sp. strain Acii25. (B) A mixture of AFV6, AFV7, and AFV8 replicated in “A. convivator.” Arrows indicate two different types of virion terminal structures, namely, AFV3-like (hollow arrows) and AFV2-like (filled arrows) structures, which are enlarged in the insets. Samples were negatively stained with 3% uranyl acetate. Bars, 1 μm and 100 nm (insets in panel B).
FIG. 2.
Electron micrographs of AFV3 virion showing features of its architecture and tail structure. (A) Whole filamentous AFV3 virion. (B) AFV3 virion after treatment with 0.3% Triton X-100. (C) Power spectrum of the straight part of the AFV3 virion shown in panel B. Arrows indicate periodicities at 4.3 nm−1. (D) Horizontal slice (thickness, 0.7 nm) of the tomographic 3D reconstruction of a negatively stained virion showing three tails at the terminus, each of which is marked by an arrow. (E) Vertical slice (thickness, 0.7 nm) showing the almost round cross section of the negatively stained virion. (F) Cryo-electron micrograph of AFV3 virions, showing intact virions (arrow 1) with bent pointed ends and partially disrupted thinner viruses (arrow 2). The contrast is inverted. Bars, 500 nm (A) and 100 nm (B, D, E, and F).
FIG. 3.
Image processing of an AFV3 virion. (a) Average 2D map computed from a set of 629 experimental images of the negatively stained sample. Images were centered and aligned before being averaged. (b) Average 2D map computed from a set of 13,541 windowed, centered, and aligned images taken from cryo-EM micrographs. The contrast is inverted so that proteins and other biomolecules appear white on a dark background. Arrows indicate the dimensions cited in the text. Bars, 5 nm.
FIG. 4.
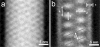
(a to c) Average 2D maps computed from three homogeneous classes obtained after correspondence analysis and hierarchical ascendant classification. The classification reveals primarily strong density variations in the lumen of the tube relating to the disposition of ovoid masses. (g) Front view of a model composed of two antiparallel rows of hollow disks simulating the ovoid masses in panels a to c, with a cylinder corresponding to the outer virus shell. (h and i) Front views of the model horizontally tilted +15° and −15°, respectively. (d to f) Projection 2D maps computed from volumes (g to i, respectively) which resemble the patterns observed in panels a, b, and c, respectively.
FIG. 5.
Genome maps aligned for AFV3, AFV6, AFV7, and AFV8 and a reannotated version of SIFV (1, 27). The maps show the relative sizes and orientations of the predicted genes, where those above the horizontal line are transcribed from left to right and those below are transcribed from right to left. Homologous genes are coded with identical colors and shading. Homologous operons carrying three or more genes are shown with graduated color coding. White boxes indicate that no homologous genes were detected in the other genomes. The predicted AFV3 ORFs are labeled according to their amino acid lengths. Predicted protein functions are shown as follows: hc, helicase; gt, glycosyltransferase; mt, SAM-dependent methyltransferase; nc, nuclease; pp, protein phosphatase; sp, structural protein; and tr, transcriptional regulator. Those ORFs which give matches with crenarchaeal chromosomal CRISPR sequences are shown in bold.
FIG. 6.
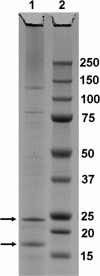
SDS-polyacrylamide gel electrophoresis of AFV3 proteins. Lane 1, viral proteins; lane 2, standard protein size markers. The proteins indicated by arrows were identified by mass spectroscopy as products of ORF166 and ORF204.
FIG. 7.
Highly conserved genomic region carrying two adjacent operons. The operon to the right encodes the two major structural proteins (ORF166 and ORF204) that may be regulated by ORF97, which shares a TATA-like sequence (AAATATAAAA) with the operon. Arrows indicate the direction of transcription. In the operon to the left, there is a 7-bp overlap of the two genes. Both operons are predicted to produce leaderless transcripts, with exceptionally strong (8 bp) predicted Shine-Dalgarno interactions for the second gene of each operon.
FIG. 8.
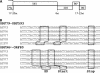
(A) Scheme showing the degree of ORF overlap in the operon carrying the putative replication protein genes (ORF593 and ORF203) of the AFV genomes. In AFV3, exceptionally, the start codon of ORF96* is mutated to ACG. (B) Sequences are aligned at the large gene overlaps shown in panel A, with Shine-Dalgarno motifs (SD) and start and stop codons shown in boxes.
FIG. 9.
(A) Large section of AFV7 considered to have resulted from intergenomic recombination at sites 1 and 2 with a 13-kb region of an AFV2-like deltalipothrixviral genome. Gene maps of this AFV2-like section are aligned with the corresponding region of AFV2, where the homologous genes are color coded (13). ORF329 of AFV7 corresponds to a glycosyltransferase. (B) Sequence alignments for AFV7 and AFV3 at the putative recombination sites 1 and 2 of the genomic fragment reveal sequence discontinuities at the two positions. Highly conserved sequences are shaded, and identical nucleotides are indicated by asterisks. Putative start codons are underlined.
FIG. 10.
(A) Sequence alignments showing examples of putative 12-bp elements and a 24-bp region located in highly conserved regions of ORF185, ORF203, and ORF1349 (AFV3 numbers). Identical nucleotides are marked with asterisks. (B) Sequence alignment of a conserved viral sequence with a CRISPR sequence in S. solfataricus P2 (17), where asterisks indicate identical nucleotides in all the viruses. Below the alignment are putative target sites on the overlapping ORFs showing that the sequence is similar to regions of both ORF96 and ORF80.
FIG. 11.
Dendrogram for the betalipothrixviruses, the gammalipothrixvirus AFV1, and the deltalipothrixvirus AFV2, derived using MEGA3 (16), for homologs of AFV3 ORF235, which is the only well-conserved ORF shared by these viruses. Schematic models of the virion structures of AFV3, AFV1, and AFV2 are superimposed on the dendrogram. Five hundred bootstrap replications were performed.
Similar articles
- AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus.
Bettstetter M, Peng X, Garrett RA, Prangishvili D. Bettstetter M, et al. Virology. 2003 Oct 10;315(1):68-79. doi: 10.1016/s0042-6822(03)00481-1. Virology. 2003. PMID: 14592760 - Structure and genome organization of AFV2, a novel archaeal lipothrixvirus with unusual terminal and core structures.
Häring M, Vestergaard G, Brügger K, Rachel R, Garrett RA, Prangishvili D. Häring M, et al. J Bacteriol. 2005 Jun;187(11):3855-8. doi: 10.1128/JB.187.11.3855-3858.2005. J Bacteriol. 2005. PMID: 15901711 Free PMC article. - A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus.
Vestergaard G, Häring M, Peng X, Rachel R, Garrett RA, Prangishvili D. Vestergaard G, et al. Virology. 2005 May 25;336(1):83-92. doi: 10.1016/j.virol.2005.02.025. Virology. 2005. PMID: 15866073 - Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses.
Prangishvili D, Garrett RA. Prangishvili D, et al. Biochem Soc Trans. 2004 Apr;32(Pt 2):204-8. doi: 10.1042/bst0320204. Biochem Soc Trans. 2004. PMID: 15046572 Review. - Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus.
Lipps G. Lipps G. Extremophiles. 2006 Feb;10(1):17-28. doi: 10.1007/s00792-005-0492-x. Epub 2006 Jan 6. Extremophiles. 2006. PMID: 16397749 Review.
Cited by
- Regulatory sequence-based discovery of anti-defense genes in archaeal viruses.
Bhoobalan-Chitty Y, Xu S, Martinez-Alvarez L, Karamycheva S, Makarova KS, Koonin EV, Peng X. Bhoobalan-Chitty Y, et al. Nat Commun. 2024 May 2;15(1):3699. doi: 10.1038/s41467-024-48074-x. Nat Commun. 2024. PMID: 38698035 Free PMC article. - DeepTracer-ID: De novo protein identification from cryo-EM maps.
Chang L, Wang F, Connolly K, Meng H, Su Z, Cvirkaite-Krupovic V, Krupovic M, Egelman EH, Si D. Chang L, et al. Biophys J. 2022 Aug 2;121(15):2840-2848. doi: 10.1016/j.bpj.2022.06.025. Epub 2022 Jun 28. Biophys J. 2022. PMID: 35769006 Free PMC article. - Imaging Techniques for Detecting Prokaryotic Viruses in Environmental Samples.
Turzynski V, Monsees I, Moraru C, Probst AJ. Turzynski V, et al. Viruses. 2021 Oct 21;13(11):2126. doi: 10.3390/v13112126. Viruses. 2021. PMID: 34834933 Free PMC article. Review. - The viral capsid as novel nanomaterials for drug delivery.
Aljabali AA, Hassan SS, Pabari RM, Shahcheraghi SH, Mishra V, Charbe NB, Chellappan DK, Dureja H, Gupta G, Almutary AG, Alnuqaydan AM, Verma SK, Panda PK, Mishra YK, Serrano-Aroca Á, Dua K, Uversky VN, Redwan EM, Bahar B, Bhatia A, Negi P, Goyal R, McCarron P, Bakshi HA, Tambuwala MM. Aljabali AA, et al. Future Sci OA. 2021 Jul 14;7(9):FSO744. doi: 10.2144/fsoa-2021-0031. eCollection 2021 Oct. Future Sci OA. 2021. PMID: 34737885 Free PMC article. Review. - A filamentous archaeal virus is enveloped inside the cell and released through pyramidal portals.
Baquero DP, Gazi AD, Sachse M, Liu J, Schmitt C, Moya-Nilges M, Schouten S, Prangishvili D, Krupovic M. Baquero DP, et al. Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2105540118. doi: 10.1073/pnas.2105540118. Proc Natl Acad Sci U S A. 2021. PMID: 34341107 Free PMC article.
References
- Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. Kristjansson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267252-266. - PubMed
- Baranov, P. V., O. Fayet, R. W. Hendrix, and J. Atkins. 2006. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 22174-181. - PubMed
- Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. - PubMed
- Benzecri, J. P., and S. Watanabe (ed.). 1969. Methodologies of pattern recognition. Academic Press, New York, NY.
- Bettstetter, M., X. Peng, R. A. Garrett, and D. Prangishvili. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 31568-79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources