Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila - PubMed (original) (raw)
Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila
Julide Bilen et al. PLoS Genet. 2007 Oct.
Abstract
Spinocerebellar ataxia type-3 (SCA3) is among the most common dominantly inherited ataxias, and is one of nine devastating human neurodegenerative diseases caused by the expansion of a CAG repeat encoding glutamine within the gene. The polyglutamine domain confers toxicity on the protein Ataxin-3 leading to neuronal dysfunction and loss. Although modifiers of polyglutamine toxicity have been identified, little is known concerning how the modifiers function mechanistically to affect toxicity. To reveal insight into spinocerebellar ataxia type-3, we performed a genetic screen in Drosophila with pathogenic Ataxin-3-induced neurodegeneration and identified 25 modifiers defining 18 genes. Despite a variety of predicted molecular activities, biological analysis indicated that the modifiers affected protein misfolding. Detailed mechanistic studies revealed that some modifiers affected protein accumulation in a manner dependent on the proteasome, whereas others affected autophagy. Select modifiers of Ataxin-3 also affected tau, revealing common pathways between degeneration due to distinct human neurotoxic proteins. These findings provide new insight into molecular pathways of polyQ toxicity, defining novel targets for promoting neuronal survival in human neurodegenerative disease.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
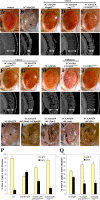
Figure 1. Modifiers of SCA3trQ78-Induced Neurodegeneration in Drosophila Eye and Internal Retinal Sections of 1-d Flies
(A) Control with normal eye and retinal thickness (white arrow). Genotype w; gmr-GAL4/+. (B) Expression of strong SCA3trQ78 causes external and internal degeneration, with pigment loss and severely reduced retinal thickness. Genotype w; gmr-GAL4 UAS-SCA3trQ78/+. (C–E) Class 1 suppressors Hsp68E407, mrjE1050, and CG14207EP1348 rescued external eye pigmentation and restored the internal eye towards normal. Genotypes w; gmr-GAL4 UAS-SCA3trQ78 in trans to alleles indicated. (F and G) Class 3 suppressors embE128-1A and dbrEP9 have strong and weak rescue. Genotypes w; gmr-GAL4 UAS-SCA3trQ78 in trans to alleles indicated. (H–J) Co-expression of CG11033EP3093 with SCA3trQ61 severely enhances pigment loss and internal retinal degeneration; expression of CG11033EP3093 alone has no effect. Genotypes (H) w; gmr-GAL4 UAS-SCA3trQ61/+, (I) w; gmr-GAL4 UAS-SCA3trQ61 in trans to CG11033EP3093, and (J) w; gmr-GAL4 in trans to CG11033EP3093. (K–O) Deletions for regions that uncover select modifier genes enhance Ataxin-3 degeneration, suggesting dose sensitivity of these modifiers. (K) Pathogenic Ataxin-3 causes neuronal degeneration. (L) Ataxin-3 in trans to Df(3L)CH20, which uncovers both DnaJ-1 and Ubp64E, is lethal and causes severe degeneration (pupal eye shown). (M–O) Ataxin-3 in trans to deficiencies uncovering Tpr2 (Df(2L)r10), emb (Df(2L)TE29A), or dbr (Df(2L)PM44). Genotypes (K) w; gmr-GAL4 UAS-SCA3trQ78/+, (L) w; gmr-GAL4 UAS-SCA3trQ78 /+; Df(3L)CH10/+, (M) w;gmr-GAL4 UAS-SCA3trQ78/ Df(2L)r10, (N) w;gmr-GAL4 UAS-SCA3trQ78/ Df(2L)TE29A, and (O) w;gmr-GAL4 UAS-SCA3trQ78/ Df(2L)PM44. (P and Q) Modifiers functionally restore vision. (P) Flies with normal vision chose light when given a choice between light and dark chambers; flies expressing SCA3tr-Q78 are blind, choosing light and dark in equal numbers (1-d flies). Co-expression of mrjE1050 or CG14207EP1348 with SCA3trQ78 restored vision toward normal. Genotypes: Canton-S, w; gmr-GAL4 UAS-SCA3trQ78 in trans to +, mrjE1050 or CG14207EP1348. (Q) Modifiers mitigate vision loss induced by full-length pathogenic Ataxin-3. Flies expressing SCA3-Q84 have strikingly compromised vision (7-d flies). Coexpression of CG14207EP1348 or mrjE1050 suppressed visual defects and restored phototaxis. Mean ± standard error of the mean of three experiments. Genotypes w; gmr-GAL4 UAS-SCA3Q84 in trans to +, mrjE1050 or CG14207EP1348. Bar in (A), 100 μm for (A–J); bar in (K), 100 μm for (K–O).
Figure 2. EP Modifiers Strongly Affect General Protein Misfolding Defects
(A) Compromised endogenous Hsp70 via expression of a dominant negative Hsp70 transgene (UAS-Hsp70.K71E) leads to a degenerate eye. Genotype w; gmr-GAL4 UAS-Hsp70.K71E/+. (B–H) The chaperone modifiers (B) Hsp68E407 and (C) CG14207EP1348 partially rescue Hsp70.K71E. (D) An enhancer of Ataxin-3 toxicity (CG11033EP3093) also suppresses general misfolding, suggesting that Ataxin-3 toxicity and misfolding do not have identical molecular mechanisms. (E) embE2-1A, (F) Sin3AB9-E, (G) NFATEP1335, and (H) ImpEP1433 also suppress the Hsp70.K71E phenotype suggesting a role of these modifiers in protein quality control. Genotypes w; gmr-GAL4 UAS-Hsp70.K71E in trans to (B) Hsp68E407, (C) CG14207EP1348, (D) CG11033EP3093, (E) embE2-1A, (F) Sin3AB9-E, (G) NFAT EP1335, and (H) Imp EP1433. (I and J) Cochaperones DnaJ-1B345.2 and Tpr2EB7-1A are pupal lethal upon expression with Hsp70.K71E, causing more severe misfolding phenotype. Genotype w; gmr-GAL4 UAS-Hsp70.K71E in trans to EP lines (I) B345.2 and (J) EB7-1A. Bar in (A), 100 μm for (A–J). (K) Western blot indicates that the modifiers do not induce expression of Hsp70. Protein samples from the 1-d fly head, with indicated genotypes, gmr-GAL4 driver. Positive control, 7-d flies expressing UAS-SCA3trQ78.
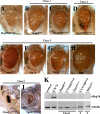
Figure 3. Modifiers Affect Accumulation or Solubility of Ataxin-3 Protein
(A–D) Horizontal cryosections of 1-d flies indicate two modes of activity of suppressors on disease protein accumulations: reduction or no effect. (A) Control flies showing NI of full-length SCA3Q84 protein. Genotype w; gmr-GAL4 UAS-SCA3Q84 /+. (B) ImpEP1433 has no effect on NI. Genotype ImpEP1433; gmr-GAL4 UAS-SCA3Q84/+. (C) Hsp68E407 and (D) CG5009B227.2 decrease NI. Genotypes (C) w; gmr-GAL4 UAS-SCA3Q84/+; E407/+ and (D) w; gmr-gal4 UAS-SCA3Q84 /B227.2. (E) Western blot shows that all suppressors increase the solubility of the disease protein. Normally, no monomer is seen in the line expressing strong SCA3trQ78 (lane 1). Upon co-expression of suppressors, monomer level of disease protein increases, indicative of increased solubility. Genotype w; gmr-GAL4 UAS-SCA3Q84 in trans to indicated suppressors. (F and H) Horizontal retinal sections of 7-d flies. (F) NI in flies expressing SCA3trQ78 are strongly reduced by Dpld. (H) Endogenous Hsp70 stress response is also strongly reduced by Dpld. Genotypes (F–H left panel) w; UAS-SCA3trQ78/+; rh1-GAL4 /+ and (F–H right panel) w; UAS-SCA3trQ78/+; rh1-GAL4 / dpld J990-P2. (I) Western blots of 7-d flies showing that reduced stress response, as determined by levels of endogenous Hsp70, correlates with increased solubility of disease protein. Dpld increases disease protein monomer level and reduces stress-induced Hsp70. Genotype w; UAS-SCA3trQ78; rh1-GAL4 in trans to +, dpldJ990-P2, dpldJM265, dpldJM265ex, and hHsp70.
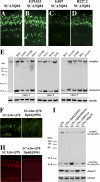
Figure 4. Degradation Pathways of Ataxin-3-Induced Neurodegeneration
(A–E) External eyes of 1-d flies showing suppression of SCAtrQ78 toxicity by modifiers only (upper panel), and suppression by modifiers in limited proteasome background (lower panel) by expression of a dominant negative form of a 20S proteasome subunit [35]. (A) Limiting proteasome activity alone does not enhance Ataxin-3 degeneration. Genotype (upper panel) w; gmr-GAL4 UAS-SCA3trQ78 /+ and (lower panel) w; gmr-GAL4 UAS-SCA3trQ78/+ ; UAS-DTS5/+. (B and C) Polyubiquitin CR11700EP1384 (B) and Ubp64EE213-1A (C) cannot suppress when proteasome activity is limited. Genotypes (B) EP1384; gmr-GAL4 UAS-SCA3trQ78 /+; UAS-DTS5/+ and (C) w; gmr-GAL4 UAS-SCA3trQ78/+; UAS-DTS5/E213-1A. (D and E) DnaJ-1B345.2 and dpldJM265 suppression remain largely unaffected when proteasome activity is limited. Genotypes (D) w; gmr-GAL4 UAS-SCA3trQ78/+; UAS-DTS5/ B345.2 and (E) w; gmr-GAL4 UAS-SCA3trQ78/JM265; UAS-DTS5/+. (F–I) Fat body tissues from larvae under starved or fed conditions, stained by Lysotraker Red (red) to highlight lysosomes and Hoescht (blue) for nuclei. Starvation strongly induces lysosomes (F), whereas fed animals normally have limited lysosomes (G). Ubiquitous expression of DnaJ-1 upregulates the induction of lysosomes in the fat bodies in starvation conditions (H), suggesting that DnaJ-1 may reduce NI in a disease situation by facilitating autophagy. Imp suppresses starvation-induced autophagy (I) although has no effect on NI, raising the possibility that it may modulate autophagic cell death. Genotypes (F–G) w, (H) w; da-GAL4/B345.2, and (I) EP1433;da-GAL4/+. (J and K) Scores of autophagy. Average values were obtained from at least 20 larvae per genotype. Error bar represents standard error of the mean of multiple experiments. (J) In fed conditions, pathogenic Ataxin-3 induces autophagy but normal Ataxin-3 has little effect. (K) In starvation conditions, expression of Ataxin-3, DnaJ-1, and Mrj further induce autophagy; on the contrary, Hsp70 and Imp suppress starvation-induced autophagy. (L–N) Retinal sections. (L) Flies (1 d) expressing SCA3Q84 have some loss of retinal integrity, which is mildly enhanced by reduction of (M) Atg5. (N) Reduction of atg5 activity alone has normal retinal morphology. Genotypes (L) w; gmr-GAL4 UAS-SCA3Q84/+, (M) UAS-Atg5_IR/UAS-Atg5_IR; gmr-GAL4 UAS-SCA3Q84/+, and (N) _gmr-GAL4/+; UAS_-Atg5_IR/UAS-Atg5_IR. (O) Western blot showing that reduction of Atg5 decreases Ataxin-3 monomer level, concomitant with enhanced toxicity. (P and Q) Cryosections for disease protein (1-d flies, anti-myc). Reduction of autophagy increases disease protein accumulation in the lamina (Q) compared to disease protein alone control (P). Hsc70.K71S also enhances, but it does not cause similar protein accumulations in the retina (unpublished data). Genotype in (P) is same as (L), and (Q) is same as (M). (R and S) Paraffin sections (1-d flies) showing suppression of Ataxin-3 degeneration by Dpld and that Dpld suppression is reduced by limiting autophagy. Genotypes (R) w; gmr-GAL4 UAS-SCA3trQ78 /JM265 and (S) UAS-Atg7_IR; gmr-GAL4 UAS-SCA3trQ78/JM265.
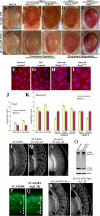
Figure 5. Select Ataxin-3 Modifiers also Modulate Tau Toxicity
(A) Top: expression of pathogenic human tau.R406W causes a rough and reduced eye [41]. Genotype w; gmr-gal4/+; UAS-tau.R406W/+. Bottom: Ectopic expression of apoptotic gene hid ablates the eye. Genotype w; gmr-GAL4 gmr-hid /+. Genes shown modulated tau.wt and tau.R406W similarly. (B–D) Tpr2EB7-1A (B), polyubiquitin CR11700EP1384 (C), and CG5009B227.2 (D) suppress pathogenic tau. Tpr2EB7-1A (B) and CR11700EP1384 (C) have no effect on Hid-induced programmed cell death. Genotypes (upper panel B) w; gmr-GAL4/+; UAS-tau.R406W /EB7-1A, (C) EP1384; gmr-GAL4/+; UAS-tau.R406W /+, and w; gmr-gal4/B227.2; UAS-tau.R406W /+. (Lower panel, B) w; gmr-GAL4 gmr-hid /EB7-1A, (C) EP1384; gmr-GAL4 gmr-hid /+, and (D) w; gmr-GAL4 gmr-hid/B227.2. (E–G) dpldJM265 (E), ImpEP1433 (F), and embE2-1A (G) modulate tau toxicity and programmed cell death in a similar manner, suggesting that their effect on tau may be due to modulation of cell death. Genotypes (upper panel, E) w; gmr-GAL4/+; UAS-tau.R406W /JM265, (F) EP1433; gmr-GAL4/+; UAS-tau.R406W /+, and (G) w; gmr-GAL4/E2-1A; UAS-tau.R406W /+. (Lower panel, E) w; gmr-GAL4 gmr-hid /JM265, (F) EP1433; gmr-GAL4 gmr-hid /+, and (G) w; gmr-GAL4 gmr-hid/E2-1A.
Figure 6. Activities of Modifier Genes of Ataxin-3-Associated Neurodegeneration
(A) Modifiers of pathogenic Ataxin-3 toxicity may (1) reduce disease protein accumulation into NI in a manner sensitive to proteasome activity and/or by modulating autophagy, (2) promote cellular functionality in situations of misfolded protein, and/or (3) promote neuronal survival by regulating autophagic cell loss. (B) Select modifiers of Ataxin-3 toxicity reveal genetic links to tau toxicity and Hid-induced programmed cell death. Tpr2EB7-1A, polyubiquitin CR11700EP1384, and CG5009B227.2 suppress both Ataxin-3 and tau toxicity, implicating chaperone and ubiquitin proteasome activity as modifiers of tau effects. Modifiers with similar effects on tau and hid-induced cell death may be modulating tau by modulating cell death; however, these modifiers likely act in a distinct way on Ataxin-3-associated degeneration, which is not sensitive to programmed cell death genes but rather appears modulated by the microRNA bantam [22].
Similar articles
- RNA toxicity is a component of ataxin-3 degeneration in Drosophila.
Li LB, Yu Z, Teng X, Bonini NM. Li LB, et al. Nature. 2008 Jun 19;453(7198):1107-11. doi: 10.1038/nature06909. Epub 2008 Apr 30. Nature. 2008. PMID: 18449188 Free PMC article. - MicroRNA pathways modulate polyglutamine-induced neurodegeneration.
Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. Bilen J, et al. Mol Cell. 2006 Oct 6;24(1):157-63. doi: 10.1016/j.molcel.2006.07.030. Mol Cell. 2006. PMID: 17018300 - Druggable genome screen identifies new regulators of the abundance and toxicity of ATXN3, the Spinocerebellar Ataxia type 3 disease protein.
Ashraf NS, Sutton JR, Yang Y, Ranxhi B, Libohova K, Shaw ED, Barget AJ, Todi SV, Paulson HL, Costa MDC. Ashraf NS, et al. Neurobiol Dis. 2020 Apr;137:104697. doi: 10.1016/j.nbd.2019.104697. Epub 2019 Nov 26. Neurobiol Dis. 2020. PMID: 31783119 Free PMC article. - Progress in pathogenesis studies of spinocerebellar ataxia type 1.
Cummings CJ, Orr HT, Zoghbi HY. Cummings CJ, et al. Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1079-81. doi: 10.1098/rstb.1999.0462. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434309 Free PMC article. Review. - Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance.
Li X, Liu H, Fischhaber PL, Tang TS. Li X, et al. Prog Neurobiol. 2015 Sep;132:34-58. doi: 10.1016/j.pneurobio.2015.06.004. Epub 2015 Jun 27. Prog Neurobiol. 2015. PMID: 26123252 Review.
Cited by
- Virus recognition by Toll-7 activates antiviral autophagy in Drosophila.
Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Nakamoto M, et al. Immunity. 2012 Apr 20;36(4):658-67. doi: 10.1016/j.immuni.2012.03.003. Epub 2012 Mar 29. Immunity. 2012. PMID: 22464169 Free PMC article. - Model systems of protein-misfolding diseases reveal chaperone modifiers of proteotoxicity.
Brehme M, Voisine C. Brehme M, et al. Dis Model Mech. 2016 Aug 1;9(8):823-38. doi: 10.1242/dmm.024703. Dis Model Mech. 2016. PMID: 27491084 Free PMC article. Review. - Modifier pathways in polyglutamine (PolyQ) diseases: from genetic screens to drug targets.
Costa MD, Maciel P. Costa MD, et al. Cell Mol Life Sci. 2022 May 3;79(5):274. doi: 10.1007/s00018-022-04280-8. Cell Mol Life Sci. 2022. PMID: 35503478 Free PMC article. Review. - Protein homeostasis in models of aging and age-related conformational disease.
Kikis EA, Gidalevitz T, Morimoto RI. Kikis EA, et al. Adv Exp Med Biol. 2010;694:138-59. doi: 10.1007/978-1-4419-7002-2_11. Adv Exp Med Biol. 2010. PMID: 20886762 Free PMC article. Review. - Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster.
Iyer J, Singh MD, Jensen M, Patel P, Pizzo L, Huber E, Koerselman H, Weiner AT, Lepanto P, Vadodaria K, Kubina A, Wang Q, Talbert A, Yennawar S, Badano J, Manak JR, Rolls MM, Krishnan A, Girirajan S. Iyer J, et al. Nat Commun. 2018 Jun 29;9(1):2548. doi: 10.1038/s41467-018-04882-6. Nat Commun. 2018. PMID: 29959322 Free PMC article.
References
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. - PubMed
- Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. - PubMed
- Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, et al. Expanded polyglutamine in the Machado-Joseph disease protein induces cell death in vitro and in vivo. Nat Genet. 1996;13:196–202. - PubMed
- Perutz MF. Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem Sci. 1999;24:58–63. - PubMed
- Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous