Novel cargo-binding site in the beta and delta subunits of coatomer - PubMed (original) (raw)
Novel cargo-binding site in the beta and delta subunits of coatomer
Kai Michelsen et al. J Cell Biol. 2007.
Abstract
Arginine (R)-based ER localization signals are sorting motifs that confer transient ER localization to unassembled subunits of multimeric membrane proteins. The COPI vesicle coat binds R-based signals but the molecular details remain unknown. Here, we use reporter membrane proteins based on the proteolipid Pmp2 fused to GFP and allele swapping of COPI subunits to map the recognition site for R-based signals. We show that two highly conserved stretches--in the beta- and delta-COPI subunits--are required to maintain Pmp2GFP reporters exposing R-based signals in the ER. Combining a deletion of 21 residues in delta-COP together with the mutation of three residues in beta-COP gave rise to a COPI coat that had lost its ability to recognize R-based signals, whilst the recognition of C-terminal di-lysine signals remained unimpaired. A homology model of the COPI trunk domain illustrates the recognition of R-based signals by COPI.
Figures
Figure 1.
Identification of regions required for the interaction with R-based signals in the δ- and β-COP subunit. (A) Yeast two-hybrid analysis of Gal4 BD fusions of five COPI subunits and a strong (R; KLRRRRI) or weak (M; NVRNRRK) R-based signal fused to the Gal4 AD performed at low stringency. Except for β-COP (yeast SEC26) all open reading frames encoding COPI subunits were bovine (Faulstich et al., 1996). (B) C-terminal truncation and internal deletion analysis of bovine δ-COP using the yeast two-hybrid assay. (C) Alignment of the critical region as mapped in bovine δ-COP with the corresponding regions of yeast, human, and fly δ-COP proteins. (D) N-terminal truncation and internal deletion analysis of β-COP using the yeast two-hybrid assay. (E) Alignment of the critical region as mapped in yeast β-COP with the corresponding regions of human, mouse, and fly β-COP proteins.
Figure 2.
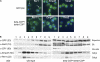
Specific loss of R-based signal-dependent sorting in cells with mutant coatomer. (A) Localization of the Pmp2YFP reporter protein presenting a strong (KLRRRRI) or a weak (NVRNRRK) R-based signal in wild-type and cells expressing mutant β-COP* (Sec26 334DLD/NAN), δ-COP* (Ret2Δ414-435), or both. Cells were grown at 25°C. Top panels: YFP fluorescence, bottom panels: DIC images. Bar, 5 μm. White arrows mark the vacuole. (B) Yeast mutants coexpressing expressing β-COP* and δ-COP* (Sec26 334DLD/NAN and Ret2Δ414-435; see methods for details of strain construction) and either Pmp2GFP-LRKR or Pmp2GFP-KKTN were stained with FM4-64 as described by Vida and Emr (1995). Top panels: GFP fluorescence, bottom panels: FM4-64 images. Bar, 5 μm. White arrow marks the vacuole. (C) Binding of wild-type (wt) and double-mutant (**) coatomer from yeast cytosol to GST fusion proteins of the cytosolic tail of Mst27 (Sandmann et al., 2003) presenting the indicated sorting signal (KKTE or LRKR). The bound fraction was eluted from the beads and analyzed by SDS PAGE and Western blotting. Blots were probed using an antiserum recognizing all COPI subunits (bands from top to bottom: α-COP/Ret1, an unresolved triplet of β-/Sec26, β′/Sec27, and γ-COP/Sec21, and δ-COP/Ret2; compare Sandmann et al. (2003) for a characterization of the antibodies) and subsequently with an antiserum recognizing the GST-fusion protein immobilized on the glutathione agarose (labeled GST fusions). The signal of the fluorescent secondary antibody was quantified (one representative blot, left). The bar diagram (right) shows the quantification of the α-COP band for seven independent experiments. The intensity of the α-COP signal was normalized to the intensity of the corresponding anti-GST signal to correct for differences in the amount of protein-loaded beads. For each cytosolic tail, the α-COP signal obtained in the pull-down was expressed as a fraction of the signal obtained from the wild-type coat.
Figure 3.
COPI-dependent retrograde cargo is correctly localized in cells with mutant coatomer supporting the specificity of the sorting defect for R-based signals. (A) Indirect immunofluorescence of the cells coexpressing β-COP* and δ-COP* (Sec26 334DLD/NAN and Ret2Δ414-435) or wild-type cells using primary antibodies against the indicated proteins and a fluorescent secondary antibody (depicted in green). Nuclei were stained with DAPI (blue). White arrows mark Rer1-positive puncta. (B) Subcellular fractionation of wild-type or double-mutant yeast cells expressing Pmp2YFP-NVRNRRK. Cells were grown at 25°C. Homogenates were prepared from spheroplasts and loaded on ten-step sucrose gradients (fraction 1: 18% sucrose, fraction 9: 50% sucrose). Membranes were sedimented from the fractions indicated and 5 μg of membrane protein was loaded in each lane followed by Western blotting analysis using the indicated antibodies (the anti-GFP signal reflects the distribution of the Pmp2YFP-NVRNRRK reporter in the gradient, Kar2 is an ER marker, CPY is a vacuolar protein, Emp47 and Rer1 are proteins that depend on COPI for their correct localization to the Golgi).
Figure 4.
Position of the identified binding site(s) in a homology model of the COPI trunk domain. (A) Stereo pairs of two different orientations of the homology model lacking the appendage domains. β-COP is shown in green, γ-COP in gray, δ-COP in light blue and ζ-COP in brown. The mapped, critical region is shown in dark blue (β-COP) and in red (δ-COP). The position of the PI4-P binding site present in the AP-1 structure is indicated by the PI4-P molecule in ball-and-stick representation. (B) Ribbon presentation of the clathrin adaptor 1 core based on the coordinates deposited by Heldwein et al. (2004). The black ellipse indicates the binding site for YXXΦ signals as shown in Fig. 1 A of Heldwein et al. (2004).
Similar articles
- Structural basis for the binding of tryptophan-based motifs by δ-COP.
Suckling RJ, Poon PP, Travis SM, Majoul IV, Hughson FM, Evans PR, Duden R, Owen DJ. Suckling RJ, et al. Proc Natl Acad Sci U S A. 2015 Nov 17;112(46):14242-7. doi: 10.1073/pnas.1506186112. Epub 2015 Nov 2. Proc Natl Acad Sci U S A. 2015. PMID: 26578768 Free PMC article. - Molecular basis for recognition of dilysine trafficking motifs by COPI.
Jackson LP, Lewis M, Kent HM, Edeling MA, Evans PR, Duden R, Owen DJ. Jackson LP, et al. Dev Cell. 2012 Dec 11;23(6):1255-62. doi: 10.1016/j.devcel.2012.10.017. Epub 2012 Nov 21. Dev Cell. 2012. PMID: 23177648 Free PMC article. - Sorting signals in the cytosolic tail of membrane proteins involved in the interaction with plant ARF1 and coatomer.
Contreras I, Ortiz-Zapater E, Aniento F. Contreras I, et al. Plant J. 2004 May;38(4):685-98. doi: 10.1111/j.1365-313X.2004.02075.x. Plant J. 2004. PMID: 15125774 - Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins.
Michelsen K, Yuan H, Schwappach B. Michelsen K, et al. EMBO Rep. 2005 Aug;6(8):717-22. doi: 10.1038/sj.embor.7400480. EMBO Rep. 2005. PMID: 16065065 Free PMC article. Review. - COPI budding within the Golgi stack.
Popoff V, Adolf F, Brügger B, Wieland F. Popoff V, et al. Cold Spring Harb Perspect Biol. 2011 Nov 1;3(11):a005231. doi: 10.1101/cshperspect.a005231. Cold Spring Harb Perspect Biol. 2011. PMID: 21844168 Free PMC article. Review.
Cited by
- The promoter of filamentation (POF1) protein from Saccharomyces cerevisiae is an ATPase involved in the protein quality control process.
Costa IM, Nasser TH, Demasi M, Nascimento RM, Netto LE, Miyamoto S, Prado FM, Monteiro G. Costa IM, et al. BMC Microbiol. 2011 Dec 28;11:268. doi: 10.1186/1471-2180-11-268. BMC Microbiol. 2011. PMID: 22204397 Free PMC article. - Mutation in archain 1, a subunit of COPI coatomer complex, causes diluted coat color and Purkinje cell degeneration.
Xu X, Kedlaya R, Higuchi H, Ikeda S, Justice MJ, Setaluri V, Ikeda A. Xu X, et al. PLoS Genet. 2010 May 20;6(5):e1000956. doi: 10.1371/journal.pgen.1000956. PLoS Genet. 2010. PMID: 20502676 Free PMC article. - The cytoplasmic tail of GM3 synthase defines its subcellular localization, stability, and in vivo activity.
Uemura S, Yoshida S, Shishido F, Inokuchi J. Uemura S, et al. Mol Biol Cell. 2009 Jul;20(13):3088-100. doi: 10.1091/mbc.e08-12-1219. Epub 2009 May 6. Mol Biol Cell. 2009. PMID: 19420140 Free PMC article. - Protein sorting at the ER-Golgi interface.
Gomez-Navarro N, Miller E. Gomez-Navarro N, et al. J Cell Biol. 2016 Dec 19;215(6):769-778. doi: 10.1083/jcb.201610031. Epub 2016 Nov 30. J Cell Biol. 2016. PMID: 27903609 Free PMC article. Review. - Endoplasmic reticulum protein targeting of phospholamban: a common role for an N-terminal di-arginine motif in ER retention?
Sharma P, Ignatchenko V, Grace K, Ursprung C, Kislinger T, Gramolini AO. Sharma P, et al. PLoS One. 2010 Jul 9;5(7):e11496. doi: 10.1371/journal.pone.0011496. PLoS One. 2010. PMID: 20634894 Free PMC article.
References
- Andag, U., T. Neumann, and H.D. Schmitt. 2001. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 276:39150–39160. - PubMed
- Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22:195–201. - PubMed
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1997. Current Protocols in Molecular Biology. Published by Greene Pub. Associates and Wiley-Interscience: J. Wiley, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases