Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta - PubMed (original) (raw)
Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta
Joel R McNeal et al. BMC Plant Biol. 2007.
Abstract
Background: Plastid genome content and protein sequence are highly conserved across land plants and their closest algal relatives. Parasitic plants, which obtain some or all of their nutrition through an attachment to a host plant, are often a striking exception. Heterotrophy can lead to relaxed constraint on some plastid genes or even total gene loss. We sequenced plastid genomes of two species in the parasitic genus Cuscuta along with a non-parasitic relative, Ipomoea purpurea, to investigate changes in the plastid genome that may result from transition to the parasitic lifestyle.
Results: Aside from loss of all ndh genes, Cuscuta exaltata retains photosynthetic and photorespiratory genes that evolve under strong selective constraint. Cuscuta obtusiflora has incurred substantially more change to its plastid genome, including loss of all genes for the plastid-encoded RNA polymerase. Despite extensive change in gene content and greatly increased rate of overall nucleotide substitution, C. obtusiflora also retains all photosynthetic and photorespiratory genes with only one minor exception.
Conclusion: Although Epifagus virginiana, the only other parasitic plant with its plastid genome sequenced to date, has lost a largely overlapping set of transfer-RNA and ribosomal genes as Cuscuta, it has lost all genes related to photosynthesis and maintains a set of genes which are among the most divergent in Cuscuta. Analyses demonstrate photosynthetic genes are under the highest constraint of any genes within the plastid genomes of Cuscuta, indicating a function involving RuBisCo and electron transport through photosystems is still the primary reason for retention of the plastid genome in these species.
Figures
Figure 1
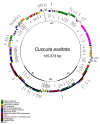
Circular map of the complete plastid genome of Ipomoea purpurea. The genome comprises an 88,172 bp LSC, a 12,110 bp SSC, and two 30,882 bp IRs. Position one of the annotated sequence begins at the LSC/IRA junction and increases numerically counterclockwise around the genome. Genes on the inside of the circle are transcribed clockwise, those on the outside, counterclockwise. Asterisks mark genes with introns (2 asterisks mark genes with 2 introns), Ψ indicates a pseudogene. INSET-Genomes scaled to relative size: Ipomoea (outermost), Cuscuta exaltata (middle), and C. obtusiflora (innermost).
Figure 2
Circular map of the complete plastid genome of Cuscuta exaltata. The genome comprises an 82,721 bp LSC and a 9,250 bp SSC separated by two 16,701 bp IRs. Inversion end-points are shown with lines connecting the inner circle to the outer. Position one of the annotated sequence begins at the LSC/IRA junction and increases numerically counterclockwise around the genome. Genes are denoted as in Figure 1.
Figure 3
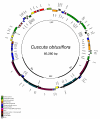
Circular map of the complete plastid genome of Cuscuta obtusiflora. The genome comprises a 50,201 bp LSC and a 6,817 bp SSC separated by 14,131 bp IRs. Position one of the annotated sequence begins at the LSC/IRA junction and increases numerically counterclockwise around the genome. Genes are denoted as in Figure 1.
Figure 4
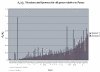
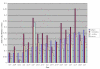
Pairwise d N/d S of Nicotiana and Ipomoea vs. Panax ginseng for all shared protein-coding genes. Genes are ranked left to right by increasing d N/d S for Nicotiana. Genes lost in Cuscuta exaltata and C. obtusiflora are indicated below the graph.
Figure 5
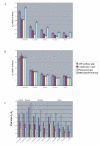
Rates of substitution and selection across 4 functionally-defined classes of genes. A- d N estimates and standard errors vs Panax for Atropa, Nicotiana, Ipomoea, C. exaltata, and C. obtusiflora. B-d S vs Panax for the same taxa. C. Pairwise d N/d S for the same taxa vs. Panax; Ipomoea, C. exaltata, and C. obtusiflora vs. Nicotiana; C. exaltata and C. obtusiflora vs. Ipomoea, and C. exaltata vs. C. obtusiflora.
Figure 6
Phylogenetic trees created using Maximum Likelihood GTR+gamma for each functionally defined gene class. Branches with significantly higher (LRT, p < 0.01) rates of synonymous substitution per site are thickened. Branches with significantly higher d N/d S are marked with one (p < 0.01), two, (p < 0.001), or three asterisks (p < 0.0001). Values of d S and d N/d S on relevant branches are given in Table 3.
Figure 7
Amino acid _p_-distance for Epifagus, Ipomoea, C. exaltata, and C. obtusiflora vs.Panax across most genes present in Epifagus. Most genes are less altered relative to the outgroup in Epifagus than in Cuscuta obtusiflora, and the non-transcriptional/translational genes remaining in Epifagus (clpP and accD) are particularly divergent in C. obtusiflora.
Figure 8
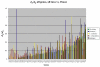
d N/d S for all genes, all taxa (including Epifagus) vs.Panax. Most genes evolve more quickly in Ipomoea than in Nicotiana (tobacco), indicating relaxed constraint on plastid genes even before evolution of parasitism in Convolvulaceae. Constraint is further relaxed in Cuscuta exaltata and is most relaxed in Cuscuta obtusiflora, although photosynthetically related genes remain highly constrained. In general, genes present in Epifagus virginiana are under higher levels of constraint than in Cuscuta obtusiflora, despite the retention of photosynthetic genes in Cuscuta.
Similar articles
- Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii.
Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Funk HT, et al. BMC Plant Biol. 2007 Aug 22;7:45. doi: 10.1186/1471-2229-7-45. BMC Plant Biol. 2007. PMID: 17714582 Free PMC article. - Parallel loss of plastid introns and their maturase in the genus Cuscuta.
McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. McNeal JR, et al. PLoS One. 2009 Jun 19;4(6):e5982. doi: 10.1371/journal.pone.0005982. PLoS One. 2009. PMID: 19543388 Free PMC article. - Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta.
Revill MJ, Stanley S, Hibberd JM. Revill MJ, et al. J Exp Bot. 2005 Sep;56(419):2477-86. doi: 10.1093/jxb/eri240. Epub 2005 Aug 1. J Exp Bot. 2005. PMID: 16061507 - Piecing together the puzzle of parasitic plant plastome evolution.
Krause K. Krause K. Planta. 2011 Oct;234(4):647-56. doi: 10.1007/s00425-011-1494-9. Epub 2011 Aug 18. Planta. 2011. PMID: 21850456 Review. - From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants.
Krause K. Krause K. Curr Genet. 2008 Sep;54(3):111-21. doi: 10.1007/s00294-008-0208-8. Epub 2008 Aug 12. Curr Genet. 2008. PMID: 18696071 Review.
Cited by
- Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants.
Wicke S, Müller KF, dePamphilis CW, Quandt D, Bellot S, Schneeweiss GM. Wicke S, et al. Proc Natl Acad Sci U S A. 2016 Aug 9;113(32):9045-50. doi: 10.1073/pnas.1607576113. Epub 2016 Jul 22. Proc Natl Acad Sci U S A. 2016. PMID: 27450087 Free PMC article. - The dynamic evolution of mobile open reading frames in plastomes of Hymenophyllum Sm. and new insight on Hymenophyllum coreanum Nakai.
Kim HT, Kim JS. Kim HT, et al. Sci Rep. 2020 Jul 6;10(1):11059. doi: 10.1038/s41598-020-68000-7. Sci Rep. 2020. PMID: 32632087 Free PMC article. - Complete Chloroplast Genomes of 14 Subspecies of D. glomerata: Phylogenetic and Comparative Genomic Analyses.
Jiao Y, Feng G, Huang L, Nie G, Li Z, Peng Y, Li D, Xiong Y, Hu Z, Zhang X. Jiao Y, et al. Genes (Basel). 2022 Sep 9;13(9):1621. doi: 10.3390/genes13091621. Genes (Basel). 2022. PMID: 36140789 Free PMC article. - Thirteen New Plastid Genomes from Mixotrophic and Autotrophic Species Provide Insights into Heterotrophy Evolution in Neottieae Orchids.
Lallemand F, Logacheva M, Le Clainche I, Bérard A, Zheleznaia E, May M, Jakalski M, Delannoy É, Le Paslier MC, Selosse MA. Lallemand F, et al. Genome Biol Evol. 2019 Sep 1;11(9):2457-2467. doi: 10.1093/gbe/evz170. Genome Biol Evol. 2019. PMID: 31396616 Free PMC article. - Gene Losses and Variations in Chloroplast Genome of Parasitic Plant Macrosolen and Phylogenetic Relationships within Santalales.
Nie L, Cui Y, Wu L, Zhou J, Xu Z, Li Y, Li X, Wang Y, Yao H. Nie L, et al. Int J Mol Sci. 2019 Nov 19;20(22):5812. doi: 10.3390/ijms20225812. Int J Mol Sci. 2019. PMID: 31752332 Free PMC article.
References
- Barkman TJ, McNeal JR, S. L, Coat G, Croom HB, Young ND, dePamphilis CW. Mitochondrial DNA suggests 12 origins of parasitism in angiosperms and implicates parasitic plants as vectors of horizontal gene transfer. to be submitted to Proceedings of the National Academy of Sciences of the United States of America.
- Bidartondo MI, Bruns TD. Extreme specificity in epiparasitic Monotropoideae (Ericaceae): widespread phylogenetic and geographical structure. Molecular Ecology. 2001;10:2285–2295. - PubMed
- Bidartondo MI, Redecker D, Hijri I, Wiemken A, Bruns TD, Dominguez L, Sersic A, Leake JR, Read DJ. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature. 2002;419:389–392. - PubMed
- Nickrent DL, Starr EM. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. Journal of Molecular Evolution. 1994;39:62–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases