PhylomeDB: a database for genome-wide collections of gene phylogenies - PubMed (original) (raw)
. 2008 Jan;36(Database issue):D491-6.
doi: 10.1093/nar/gkm899. Epub 2007 Oct 25.
Affiliations
- PMID: 17962297
- PMCID: PMC2238872
- DOI: 10.1093/nar/gkm899
PhylomeDB: a database for genome-wide collections of gene phylogenies
Jaime Huerta-Cepas et al. Nucleic Acids Res. 2008 Jan.
Abstract
The complete collection of evolutionary histories of all genes in a genome, also known as phylome, constitutes a valuable source of information. The reconstruction of phylomes has been previously prevented by large demands of time and computer power, but is now feasible thanks to recent developments in computers and algorithms. To provide a publicly available repository of complete phylomes that allows researchers to access and store large-scale phylogenomic analyses, we have developed PhylomeDB. PhylomeDB is a database of complete phylomes derived for different genomes within a specific taxonomic range. All phylomes in the database are built using a high-quality phylogenetic pipeline that includes evolutionary model testing and alignment trimming phases. For each genome, PhylomeDB provides the alignments, phylogentic trees and tree-based orthology predictions for every single encoded protein. The current version of PhylomeDB includes the phylomes of Human, the yeast Saccharomyces cerevisiae and the bacterium Escherichia coli, comprising a total of 32 289 seed sequences with their corresponding alignments and 172 324 phylogenetic trees. PhylomeDB can be publicly accessed at http://phylomedb.bioinfo.cipf.es.
Figures
Figure 1.
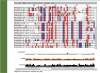
Screenshot of a PhylomeDB entry page. The entry page of an individual seed protein (Hsa0017176) of the human phylome (Hsapiens001), includes information on the sequence and links to (i) the feature page containing all necessary information of the phylogenetic pipeline used; (ii) all the phylogenetic trees derived by the different programs and models used in the pipeline, in this case a NJ tree, ML trees derived from JTT, WAG, RtREV and BLOSUM62 models, and the consensus tree derived from the Bayesian analyses; (iii) the raw and trimmed (clean) alignments and (iv) the list of tree-based orthology predictions for the species included in the tree.
Figure 3.
Visualization of phylogenetic trees: different views of the ML phylogenetic tree, using JTT evolutionary model, for the seed protein (Hsa0017176), as displayed by the environment for tree exploration tool (ETE). Trees can be represented in rectangular (A), circular (B) or radial (C) modes. Several information labels such as support values or branch distances (A) can be displayed on nodes and edges, which can also be colored to indicate different evolutionary event. In (A) an example is shown where the lineages leading to the seed protein (Hsa0017176) are colored in blue or red to mark duplications and speciation events, respectively. This feature is shown after running the orthology prediction algorithm by clicking on the ‘orthologs’ icon. Using the ETE toolbar (icons on the top of the section A), the user can navigate within the tree and browse it using different visualization options.
Figure 2.
Visualization of multiple sequence alignments: Example of the trimmed (clean) multiple sequence alignment of the example entry (Hsa0017176) as displayed by JalView (17). Sequence names are indicated by PhylomeID names, which include the three-letters species code followed by a number. The levels of sequence conservation and quality of each column, as well as a consensus sequence are indicated bellow. Colors, sequence format and visualization options can be changed using JalView interface.
Similar articles
- PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions.
Huerta-Cepas J, Capella-Gutierrez S, Pryszcz LP, Denisov I, Kormes D, Marcet-Houben M, Gabaldón T. Huerta-Cepas J, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D556-60. doi: 10.1093/nar/gkq1109. Epub 2010 Nov 12. Nucleic Acids Res. 2011. PMID: 21075798 Free PMC article. - PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome.
Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. Huerta-Cepas J, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D897-902. doi: 10.1093/nar/gkt1177. Epub 2013 Nov 25. Nucleic Acids Res. 2014. PMID: 24275491 Free PMC article. - PhylomeDB V5: an expanding repository for genome-wide catalogues of annotated gene phylogenies.
Fuentes D, Molina M, Chorostecki U, Capella-Gutiérrez S, Marcet-Houben M, Gabaldón T. Fuentes D, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D1062-D1068. doi: 10.1093/nar/gkab966. Nucleic Acids Res. 2022. PMID: 34718760 Free PMC article. - BIR Pipeline for Preparation of Phylogenomic Data.
Kumar S, Krabberød AK, Neumann RS, Michalickova K, Zhao S, Zhang X, Shalchian-Tabrizi K. Kumar S, et al. Evol Bioinform Online. 2015 Apr 27;11:79-83. doi: 10.4137/EBO.S10189. eCollection 2015. Evol Bioinform Online. 2015. PMID: 25987827 Free PMC article. Review. - The SUPERFAMILY database in structural genomics.
Gough J. Gough J. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1897-900. doi: 10.1107/s0907444902015160. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393919 Review.
Cited by
- Towards Consensus Gene Ages.
Liebeskind BJ, McWhite CD, Marcotte EM. Liebeskind BJ, et al. Genome Biol Evol. 2016 Jun 27;8(6):1812-23. doi: 10.1093/gbe/evw113. Genome Biol Evol. 2016. PMID: 27259914 Free PMC article. - BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments.
Criscuolo A, Gribaldo S. Criscuolo A, et al. BMC Evol Biol. 2010 Jul 13;10:210. doi: 10.1186/1471-2148-10-210. BMC Evol Biol. 2010. PMID: 20626897 Free PMC article. - Cross-species meta-analysis of transcriptome changes during the morula-to-blastocyst transition: metabolic and physiological changes take center stage.
Schall PZ, Latham KE. Schall PZ, et al. Am J Physiol Cell Physiol. 2021 Dec 1;321(6):C913-C931. doi: 10.1152/ajpcell.00318.2021. Epub 2021 Oct 20. Am J Physiol Cell Physiol. 2021. PMID: 34669511 Free PMC article. - Genome sequence of the pea aphid Acyrthosiphon pisum.
International Aphid Genomics Consortium. International Aphid Genomics Consortium. PLoS Biol. 2010 Feb 23;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. PLoS Biol. 2010. PMID: 20186266 Free PMC article. - Molecular characterization of the evolution of phagosomes.
Boulais J, Trost M, Landry CR, Dieckmann R, Levy ED, Soldati T, Michnick SW, Thibault P, Desjardins M. Boulais J, et al. Mol Syst Biol. 2010 Oct 19;6:423. doi: 10.1038/msb.2010.80. Mol Syst Biol. 2010. PMID: 20959821 Free PMC article.
References
- Comas I, Moya A, Gonzalez-Candelas F. From phylogenetics to phylogenomics: the evolutionary relationships of insect endosymbiotic gamma-Proteobacteria as a test case. Syst. Biol. 2007;56:1–16. - PubMed
- Gabaldón T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301:609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases