High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor - PubMed (original) (raw)
High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor
Vadim Cherezov et al. Science. 2007.
Abstract
Heterotrimeric guanine nucleotide-binding protein (G protein)-coupled receptors constitute the largest family of eukaryotic signal transduction proteins that communicate across the membrane. We report the crystal structure of a human beta2-adrenergic receptor-T4 lysozyme fusion protein bound to the partial inverse agonist carazolol at 2.4 angstrom resolution. The structure provides a high-resolution view of a human G protein-coupled receptor bound to a diffusible ligand. Ligand-binding site accessibility is enabled by the second extracellular loop, which is held out of the binding cavity by a pair of closely spaced disulfide bridges and a short helical segment within the loop. Cholesterol, a necessary component for crystallization, mediates an intriguing parallel association of receptor molecules in the crystal lattice. Although the location of carazolol in the beta2-adrenergic receptor is very similar to that of retinal in rhodopsin, structural differences in the ligand-binding site and other regions highlight the challenges in using rhodopsin as a template model for this large receptor family.
Figures
Figure 1
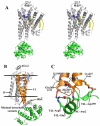
Overall fold of the β2AR-T4L fusion with its predicted orientation in the plasma membrane and key intramolecular interactions. A. Stereoview of the overall fold of β2AR-T4L. The receptor and T4L are colored gray and green, respectively. Carazolol is colored blue and the lipid molecules bound to the receptor are colored yellow. B. The receptor is aligned to a rhodopsin model that was positioned in a lipid membrane (boundaries indicated by horizontal black lines) as found in the orientations of proteins in membranes (OPM) database (74). T4L is fused internally into the third intracellular loop of β2AR and maintains minimal intramolecular packing interactions by tilting away from the receptor. C. Specific intramolecular interactions between β2AR and T4L are represented.
Figure 2
Crystal packing interactions in the lipidic mesophase crystallized β2AR-T4L. A. There are four main contact areas, two of which are mediated by T4L in the plane of the membrane with itself through a two-fold symmetry axis and translation. The third interaction is normal to the membrane plane between T4L and lumen exposed loops of β2AR. The fourth interaction is generated by the two-fold symmetry axis, packing one receptor to receptor in the plane of the membrane. B. The receptor crystal packing interface is composed mainly of lipids with two cholesterol molecules and two palmitic acid molecules forming the majority of the interactions. A network of ionic charge interactions exists on the cytoplasmic end of the interface forming the only inter-receptor protein contacts. C. Comparison between β2AR-T4L and rhodopsin (PDB ID Code 2I35) parallel receptor association interface. Helices I (blue) and VIII (magenta) are highlighted in both structures. Only one monomer is shown for each receptor representation along with helices I’ and VIII’ only from the opposing symmetry related molecule. The rhodopsin interface is twisted significantly relative to β2AR-T4L resulting in a significant offset from the parallel orientation required for a physiological dimer interface. β2AR-T4L associated monomers are in a highly parallel orientation.
Figure 3
Surface representation of β2AR colored by calculated charge from red (-10 kbT/ec) to blue (+10 kbT/ec) using a dielectric constant of 70. A. Three main areas of interest are indicated. The binding site cleft is negatively charged as is a groove between helices III, IV and V. The third region is an overall positive charge in the region of the ionic lock and DRY motif on the cytoplasmic face. The overall result is a highly polarized molecule that may utilize its negative charge to facilitate binding of catecholamine ligands. The presence of a negative charge in the groove between helices III, IV and V is unexpected as it is in the middle of the lipid membrane. This charge may be partially derived from the presence of an unpaired glutamate at position 1223.41. The effective charge in this region is likely greater than shown here due to its location in the low dielectric environment of the lipid membrane. B. View rotated 90° from A. Showing both the negatively charged binding site cleft (top) and positively charged cytoplasmic face (bottom). Poisson-Boltzmann electrostatics were calculated using the program APBS (53) as implemented in Pymol (75). Pymol was used exclusively in the preparation of all figures.
Figure 4
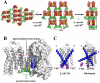
Comparison of the extracellular sides of β2AR-T4L and rhodopsin. A. The N-terminus is missing from the experimental density in the β2AR-T4L structure and is not shown. ECL2 is shown in green and contains a short α-helix and two disulfide bonds (yellow). The intraloop disulfide bond constrains the tip of ECL2 which interacts with ECL1. The second disulfide bond links ECL2 with helix III. There is one interaction between ECL2 and carazolol (blue) through Phe1935.32. The entire loop is held out of the ligand binding site by a combination of the rigid helical segment and the two disulfide bonds. B. In contrast, ECL2 (green) in rhodopsin assumes a lower position in the structure that occludes direct access to the retinal-binding site and forms a small β-sheet in combination with the N-terminal region (magenta) directly above the bound retinal (pink).
Figure 5
Ligand binding characterization and comparison to rhodopsin. A. A view looking down on the plane of the membrane from the extracellular surface showing a detailed representation of the carazolol binding site in β2AR-T4L. Carazolol is shown as sticks with carbon atoms colored yellow. β2AR-T4L residues contributing to carazolol binding are shown in green and labeled. Electron density is contoured at 5σ from an Fo-Fc omit map calculated without the contribution of carazolol. B. Binding orientation comparison between 11-_cis_-retinal in rhodopsin and carazolol in β2AR-T4L. Van der Waals’ surfaces for carazolol and retinal are represented as dots to accentuate the close packing interactions. Retinal in the all-cis conformation (pink), binds deep in the active site of rhodopsin as compared to carazolol (blue), packing its β-ionone ring between Tyr2686.51 and Phe2125.47 (cyan), blocking movement of Trp2656.48 (magenta) into the space. The β-ionone ring of _trans_-retinal in activated rhodopsin would not block Trp2656.48 from rotating into the space allowing a rotameric shift into its proposed active form. C. There are four residues involved in the toggle switch mechanism of β2AR-T4L as shown. Phe2906.52 (magenta) is sandwiched between Phe2085.47 (tan) and Phe2896.51 (tan) forming a ring-face aromatic interaction. Like rhodopsin, an activation step is thought to occur by a rotameric change of Trp2866.48 (magenta) which would displace Phe2906.52. Carazolol is shown to interact extensively with the sandwich motif as shown: however, few interactions are seen with Trp2866.48. The 6.52 position in β2AR-T4L is occupied by Phe2906.52 as opposed to Ala2696.52 in rhodopsin where the β-ionone ring replaces an aromatic protein side chain in forming the sandwich interactions. The aromatic character of the sandwich is otherwise maintained by Phe2896.51 and Phe2085.47 in β2AR-T4L.
Figure 6
Comparison of β2AR-T4L helical orientations with rhodopsin (PDB ID Code 1U19). A. β2AR-T4L is rendered as a ribbon trace colored with a blue to red spectrum corresponding to observed distances between Cα positions in the two structures (RMSD 2.7 Å between all residues in the transmembrane region). Helix II shows very little movement, whereas the entire lengths of helices III, IV, V shift significantly. Helix VIII and loops were not included in the comparison and are colored in tan. B. Movements of helices I and V of rhodopsin (grey) are shown relative to β2AR-T4L. C. Movements of helices III, IV and VI. D. Ligand binding site representation. Carazolol is shown with yellow carbons. Entire helices are assigned a single designation based on their divergence from the rhodopsin position in the area of the ligand binding site as shown. Helix I is highly divergent, Helices II and VI are similar to rhodopsin. Helices IV and VII are moderately constant. Helices III and V are moderately divergent.
Comment in
- Biochemistry. Signaling across the cell membrane.
Ranganathan R. Ranganathan R. Science. 2007 Nov 23;318(5854):1253-4. doi: 10.1126/science.1151656. Science. 2007. PMID: 18033872 No abstract available.
Similar articles
- GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function.
Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Rosenbaum DM, et al. Science. 2007 Nov 23;318(5854):1266-73. doi: 10.1126/science.1150609. Epub 2007 Oct 25. Science. 2007. PMID: 17962519 - Biochemistry. Signaling across the cell membrane.
Ranganathan R. Ranganathan R. Science. 2007 Nov 23;318(5854):1253-4. doi: 10.1126/science.1151656. Science. 2007. PMID: 18033872 No abstract available. - Crystallization of G protein-coupled receptors.
Salom D, Padayatti PS, Palczewski K. Salom D, et al. Methods Cell Biol. 2013;117:451-68. doi: 10.1016/B978-0-12-408143-7.00024-4. Methods Cell Biol. 2013. PMID: 24143992 Free PMC article. - Structural features of β2 adrenergic receptor: crystal structures and beyond.
Bang I, Choi HJ. Bang I, et al. Mol Cells. 2015;38(2):105-11. doi: 10.14348/molcells.2015.2301. Epub 2014 Dec 24. Mol Cells. 2015. PMID: 25537861 Free PMC article. Review. - G protein-coupled receptors--recent advances.
Latek D, Modzelewska A, Trzaskowski B, Palczewski K, Filipek S. Latek D, et al. Acta Biochim Pol. 2012;59(4):515-29. Epub 2012 Dec 18. Acta Biochim Pol. 2012. PMID: 23251911 Free PMC article. Review.
Cited by
- VPS26 Moonlights as a β-Arrestin-like Adapter for a 7-Transmembrane RGS Protein in Arabidopsis thaliana.
Lou F, Zhou W, Tunc-Ozdemir M, Yang J, Velazhahan V, Tate CG, Jones AM. Lou F, et al. Biochemistry. 2024 Nov 19;63(22):2990-2999. doi: 10.1021/acs.biochem.4c00361. Epub 2024 Oct 28. Biochemistry. 2024. PMID: 39467170 Free PMC article. - Active state structures of a bistable visual opsin bound to G proteins.
Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ. Tejero O, et al. Nat Commun. 2024 Oct 16;15(1):8928. doi: 10.1038/s41467-024-53208-2. Nat Commun. 2024. PMID: 39414813 Free PMC article. - Integrative residue-intuitive machine learning and MD Approach to Unveil Allosteric Site and Mechanism for β2AR.
Chen X, Wang K, Chen J, Wu C, Mao J, Song Y, Liu Y, Shao Z, Pu X. Chen X, et al. Nat Commun. 2024 Sep 16;15(1):8130. doi: 10.1038/s41467-024-52399-y. Nat Commun. 2024. PMID: 39285201 Free PMC article. - Interaction and dynamics of chemokine receptor CXCR4 binding with CXCL12 and hBD-3.
Penfield J, Zhang L. Penfield J, et al. Commun Chem. 2024 Sep 13;7(1):205. doi: 10.1038/s42004-024-01280-6. Commun Chem. 2024. PMID: 39271963 Free PMC article. - Early Events in β2AR Dimer Dynamics Mediated by Activation-Related Microswitches.
Kotipalli A, Koulgi S, Jani V, Sonavane U, Joshi R. Kotipalli A, et al. J Membr Biol. 2024 Dec;257(5-6):323-344. doi: 10.1007/s00232-024-00324-1. Epub 2024 Sep 6. J Membr Biol. 2024. PMID: 39240374
References
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. FEBS Lett. 2002;520:97. - PubMed
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. Mol Pharmacol. 2003;63:1256. - PubMed
- Pierce KL, Premont RT, Lefkowitz RJ. Nat Rev Mol Cell Biol. 2002;3:639. - PubMed
- Lefkowitz RJ, Shenoy SK. Science. 2005;308:512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- F32 GM082028/GM/NIGMS NIH HHS/United States
- P50 GM073197/GM/NIGMS NIH HHS/United States
- P50 GM062411/GM/NIGMS NIH HHS/United States
- R21 GM075811/GM/NIGMS NIH HHS/United States
- GM075915/GM/NIGMS NIH HHS/United States
- R01 NS028471/NS/NINDS NIH HHS/United States
- U54 GM074961/GM/NIGMS NIH HHS/United States
- NS028471/NS/NINDS NIH HHS/United States
- U54 GM074961-030001/GM/NIGMS NIH HHS/United States
- R01 GM089857/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- P50 GM073197-04/GM/NIGMS NIH HHS/United States
- R01 GM056169/GM/NIGMS NIH HHS/United States
- R37 NS028471/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases