Mammalian mirtron genes - PubMed (original) (raw)
Comparative Study
Mammalian mirtron genes
Eugene Berezikov et al. Mol Cell. 2007.
Abstract
Mirtrons are alternative precursors for microRNA biogenesis that were recently described in invertebrates. These short hairpin introns use splicing to bypass Drosha cleavage, which is otherwise essential for the generation of canonical animal microRNAs. Using computational and experimental strategies, we now establish that mammals have mirtrons as well. We identified 3 mirtrons that are well conserved and expressed in diverse mammals, 16 primate-specific mirtrons, and 46 candidates supported by limited cloning evidence in primates. As with some fly and worm mirtrons, the existence of well-conserved mammalian mirtrons indicates their relatively ancient incorporation into endogenous regulatory pathways. However, as worms, flies, and mammals each have different sets of mirtrons, we hypothesize that different animals may have independently evolved the capacity for this hybrid small RNA pathway. This notion is supported by our observation of several clade-specific features of mammalian and invertebrate mirtrons.
Figures
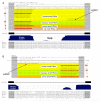
Figure 1. Examples of Mammalian Mirtrons
(A) A well-conserved mammalian mirtron. (Top) The 13th intron of the ATP-binding cassette F-1 gene harbors the mirtron mir-877. This intron is bounded by consensus splice donor and acceptor sequences, and efficient processing of this intron was evidenced by the existence of over 50 spliced cDNA clones in EST databases. The hairpin structure of this mirtron is indicated with bracket notation. Human small RNAs corresponding precisely to the 5′ and 3′ ends of the intron were identified, as were 5′ small RNAs from macaque, mouse, and rat. Cloning frequencies define the left arm product of mir-877 as its “miRNA” and the right arm product as the “miRNA*.” (Bottom) Evolutionary characteristics of this mirtron. Sequence alignment and conservation track were obtained from
. mir-877 is highly conserved among diverse eutherian species but exhibits accelerated divergence within the loop region. (B) A primate-specific mirtron. (Top) The 21st intron of the putative helicase DHX30 gene harbors the mirtron mir-1226. Notation and layout are as described in (A). In this case, cloning frequencies define its right arm product as the miRNA and its left arm product as the miRNA*. (Bottom) This mirtron is identifiable only in primates; the conservation of its 3′-most terminal sequence in other mammals likely reflects the pressure to maintain splice recognition determinants.
Figure 2. Nineteen Confidently Annotated Mammalian Mirtron Loci
These are divided into three categories: mirtrons that are conserved among diverse mammals and cloned from two or more species (three genes), mirtrons that are conserved among diverse primates and cloned from two or more species (two genes), and mirtrons that were cloned from independent libraries from a single species or three or more times from any single library (14 genes). The mirtron hairpin structures are designated with bracket notation, and exon-intron structure with “ >” and “+” notation. The cloned species are capitalized and highlighted green. Supplementary figures provide more detailed information on the cloned species (Figure S3), their tissue subtype of origin (Figure S4), and possible orthologs in other primates (Figure S5). In addition, information on 46 additional mirtron candidates is found in Figure S6.
Figure 3. Short Hairpin Introns Are the Predominant Source of Cloned Intron-Terminal Small RNAs in Diverse Mammals
Human and macaque introns were binned into 100 nt intervals. We then binned all small RNA reads derived from intron termini by intron length, excluding introns that also generated nonboundary reads (thus excluding cloned small RNAs arising from unannotated intronic noncoding RNA genes such as tRNAs or snoRNAs). It is evident that a majority of intron-terminal small RNAs in human, macaque, chimp, and mouse derive from 1–200 nt introns, and that most of these derive in turn from hairpin introns that we annotated as mirtrons or mirtron candidates.
Figure 4. Sequence and Structural Features of Mammalian and Invertebrate Mirtrons
(A) Sequence logos of 5′ and 3′ mirtron products. Data represent 19 primate/mammalian mirtrons (this study) and 18 invertebrate (14 fly and 4 worm) mirtrons (Ruby et al., 2007a). (Top row) Mammalian mirtrons generate G-rich 5′ mirtron products and C-rich 3′ mirtron products. Alignment of the 3′ mirtron products by their first nucleotides shows an equal frequency of U and C residues. (Bottom row) Invertebrate mirtrons do not show such overall G:C bias, and their 3′ products are strongly biased toward 5′ U residues. (B) Typical hairpin-end structures of mammalian and invertebrate mirtrons. These preferred end structures are also evident from the sequence logos presented in (A). (C) Comparison of the nucleotide composition of mammalian and invertebrate mirtrons with bulk short introns in humans and flies. We analyzed the GC content of 13,453 human introns and 29,120 D. melanogaster introns, each 50–120 nt in length. We also analyzed their 5′-most and 3′-most 24 nt (intron “ends”) as a proxy for miRNA/miRNA* regions. GC content and minimum free energy (mfe, kcal/mol) of straight hairpin structures for the cloned mammalian and fly mirtrons were also assessed; where only one mirtron product was obtained, the miRNA* region was inferred by assuming a 2 nt 3′ overhang. Values are shown ±SD. For comparison, we show the GC content of all human and worm/fly (invertebrate) canonical miRNAs listed in miR-base Release 10.
Similar articles
- Mirtrons: microRNA biogenesis via splicing.
Westholm JO, Lai EC. Westholm JO, et al. Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review. - Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates.
Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC. Wen J, et al. PLoS Comput Biol. 2015 Sep 1;11(9):e1004441. doi: 10.1371/journal.pcbi.1004441. eCollection 2015 Sep. PLoS Comput Biol. 2015. PMID: 26325366 Free PMC article. - RNA G-quadruplexes regulate mammalian mirtron biogenesis.
Salim U, Menon MB, Dhamija S, Vivekanandan P. Salim U, et al. J Biol Chem. 2025 Mar;301(3):108276. doi: 10.1016/j.jbc.2025.108276. Epub 2025 Feb 7. J Biol Chem. 2025. PMID: 39922486 Free PMC article. - The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila.
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Okamura K, et al. Cell. 2007 Jul 13;130(1):89-100. doi: 10.1016/j.cell.2007.06.028. Epub 2007 Jun 28. Cell. 2007. PMID: 17599402 Free PMC article. - Biogenesis, characterization, and functions of mirtrons.
Salim U, Kumar A, Kulshreshtha R, Vivekanandan P. Salim U, et al. Wiley Interdiscip Rev RNA. 2022 Jan;13(1):e1680. doi: 10.1002/wrna.1680. Epub 2021 Jun 21. Wiley Interdiscip Rev RNA. 2022. PMID: 34155810 Review.
Cited by
- The use of high-throughput sequencing methods for plant microRNA research.
Ma X, Tang Z, Qin J, Meng Y. Ma X, et al. RNA Biol. 2015;12(7):709-19. doi: 10.1080/15476286.2015.1053686. RNA Biol. 2015. PMID: 26016494 Free PMC article. Review. - Efflux ABC transporters in drug disposition and their posttranscriptional gene regulation by microRNAs.
Wang Y, Tu MJ, Yu AM. Wang Y, et al. Front Pharmacol. 2024 Jul 24;15:1423416. doi: 10.3389/fphar.2024.1423416. eCollection 2024. Front Pharmacol. 2024. PMID: 39114355 Free PMC article. Review. - microRNA involvement in human cancer.
Iorio MV, Croce CM. Iorio MV, et al. Carcinogenesis. 2012 Jun;33(6):1126-33. doi: 10.1093/carcin/bgs140. Epub 2012 Apr 3. Carcinogenesis. 2012. PMID: 22491715 Free PMC article. Review. - Deciphering microRNA code in pain and inflammation: lessons from bladder pain syndrome.
Gheinani AH, Burkhard FC, Monastyrskaya K. Gheinani AH, et al. Cell Mol Life Sci. 2013 Oct;70(20):3773-89. doi: 10.1007/s00018-013-1275-7. Epub 2013 Mar 5. Cell Mol Life Sci. 2013. PMID: 23463234 Free PMC article. Review. - Evolutionary and expression analysis of miR-#-5p and miR-#-3p at the miRNAs/isomiRs levels.
Guo L, Yu J, Yu H, Zhao Y, Chen S, Xu C, Chen F. Guo L, et al. Biomed Res Int. 2015;2015:168358. doi: 10.1155/2015/168358. Epub 2015 May 5. Biomed Res Int. 2015. PMID: 26075215 Free PMC article.
References
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006a;38:1375–1377. - PubMed
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials