Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome - PubMed (original) (raw)
Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome
Ziva Misulovin et al. Chromosoma. 2008 Feb.
Abstract
The cohesin complex is a chromosomal component required for sister chromatid cohesion that is conserved from yeast to man. The similarly conserved Nipped-B protein is needed for cohesin to bind to chromosomes. In higher organisms, Nipped-B and cohesin regulate gene expression and development by unknown mechanisms. Using chromatin immunoprecipitation, we find that Nipped-B and cohesin bind to the same sites throughout the entire non-repetitive Drosophila genome. They preferentially bind transcribed regions and overlap with RNA polymerase II. This contrasts sharply with yeast, where cohesin binds almost exclusively between genes. Differences in cohesin and Nipped-B binding between Drosophila cell lines often correlate with differences in gene expression. For example, cohesin and Nipped-B bind the Abd-B homeobox gene in cells in which it is transcribed, but not in cells in which it is silenced. They bind to the Abd-B transcription unit and downstream regulatory region and thus could regulate both transcriptional elongation and activation. We posit that transcription facilitates cohesin binding, perhaps by unfolding chromatin, and that Nipped-B then regulates gene expression by controlling cohesin dynamics. These mechanisms are likely involved in the etiology of Cornelia de Lange syndrome, in which mutation of one copy of the NIPBL gene encoding the human Nipped-B ortholog causes diverse structural and mental birth defects.
Figures
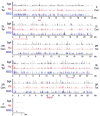
Fig. 1
Binding of the Smc1 cohesin subunit to the non-repetitive genome in Sg4, Kc, and ML-DmBG3 (BG3) Drosophila cells as determined by chromatin immunoprecipitation. The three tracks for each chromosome arm plot the trimmed mean log2 IP/control ratios for Smc1 for Sg4 (black), Kc (red) and BG3 (blue) cells on a scale from 0.5 to 2.5. The positions of the telomeres (Tel) and centromeres (Cen) are indicated for each arm. The positions of the 2-Mb region of chromosome arm 3L shown in Fig. 2, and the 4.25 Mb region plotted in Fig. 3 are indicated with arrows underneath the map, and the positions of the cut and Abd-B genes shown in detail in other figures are indicated on the X and 3R maps. These data show that the cohesin binding patterns are very similar but not identical between the three cell lines, and that with the exception of the small chromosome 4, cohesin binds many regions along the chromosome arms. There are also large cohesin-free regions extending up to a megabase or so in size, including those around 15 Mb on the map of the chromosome 2L arm and 1 Mb on the 2R map. Many large cohesin-poor regions have low gene density
Fig. 2
Binding of Nipped-B, cohesin subunits, and PolII to a 2 Mb region of chromosome 3L. This region was chosen to illustrate typical features of cohesin and Nipped-B binding patterns seen throughout the genome. The eight tracks at the top graph the trimmed mean log2 IP/control ratios for the microarray features on a scale of −0.5 to 3.0. The top four tracks (black) show RNA polymerase II (PolII), Nipped-B, and SA and Smc1 cohesin subunit binding in Sg4 cells. The red track shows Smc1 binding in Kc cells, and the three blue tracks show the PolII, Nipped-B, and Smc1 binding in BG3 cells. The vertical lines underneath the Sg4 Nipped-B, SA, and Smc1 tracks indicate microarray features predicted by TiMAT analysis to be binding peaks with a 1% false discovery rate. The annotated map of the chromosome 3L region (April 2004 release of the Drosophila melanogaster genome; Celniker et al. 2002; Berkeley Drosophila Genome Project, personal communication) is shown below the ChIP tracks. Black boxes indicate exons and lines indicate introns. The key features to note are that Nipped-B binding is virtually identical to that of the SA and Smc1 cohesin subunits, that the cohesin/Nipped-B binding patterns are very similar but not identical between the three cells lines, and that cohesin binds large regions ranging in size from a kilobase or so to more than 60 kb in length. There are also large regions, such as the 400 kb gene-poor domain near the middle of the graph that are nearly devoid of cohesin and Nipped-B
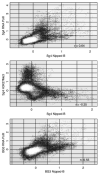
Fig. 3
Colocalization of Nipped-B and cohesin subunit binding sites. The plots compare enrichment values for the SA and Smc1 cohesin subunits and Nipped-B at individual microarray features. The trimmed mean log2 IP/control values of 1×105 microarray features for chromosome 2L extending from nt 5,522–4,254,929 (Fig. 1, chromosome 2L, 0.055 to 4.25 Mb) for the indicated proteins and cells are plotted against each other. Plots of similar-sized regions across the genome are very similar. The correlation coefficients (r) for the plotted region are given, which are similar to those calculated for the entire non-repetitive genome (Table 1). Nipped-B, SA, and Smc1 values for individual microarray features correlate well with each other except for a few Smc1-positive features that are low for SA and Nipped-B in Sg4 cells (arrows), indicating that the Nipped-B and cohesin subunit peaks closely align with each other. The white masses centered close to log2 values of 0 for both proteins represent the majority of features that have low binding for both proteins.
Fig. 4
Similarity of cohesin binding in different cell lines. The Smc1 (Sg4, Kc, and BG3) or Nipped-B (Sg4 and BG3) trimmed mean log2 IP/control values of individual microarray features for different cell lines are plotted against each other. The plots cover the same region used in Fig. 2 (chromosome 2L nt 5,522–4,254,929), and other regions show very similar results. The correlation coefficients (r) for the plotted regions are similar to those for the entire genome (Table 1). In all cases, the similarities in cohesin binding between cell lines predominate, with many features showing similar values between the two cell types being compared, indicating that that the peaks occur in precisely the same positions in the two cell lines. There are also features that have significant values in one cell type and not the other, indicating that there are also peaks specific to each cell type
Fig. 5
Binding of Nipped-B and cohesin to the cut gene regulatory region and transcription unit. Tracks above the chromosome map show cohesin subunit, Nipped-B, and PolII binding as trimmed mean log2 IP/control values (scale −0.5 to 3) for Sg4 (black), Kc (red) and BG3 (blue) cells. The peaks predicted with a 1% false discovery rate for Nipped-B, SA, and Smc1 in Sg4 cells are indicated with vertical lines underneath the tracks. The extent and direction of cut transcription is indicated with a left-to-right arrow above the cut transcript maps, and the distal wing margin enhancer is indicated by a box (wm). Nipped-B and cohesin bind to the upstream regulatory region and cut transcription unit in all three cell lines, but the binding is more extensive in BG3 cells. The promoter proximal peaks 0.5 and 4 kb upstream of the transcription start site in Kc cells (asterisks) occur precisely in the same positions as previously mapped by conventional ChIP experiments (Dorsett et al. 2005). PolII is found predominantly at the promoter in both Sg4 and BG3 cells, indicating that the difference in cohesin binding between the two cell types is unlikely to reflect a substantial difference in transcription
Fig. 6
Overlap of Nipped-B and RNA polymerase II (PolII) binding and lack of Nipped-B binding to regions enriched in histone H3 lysine 27 trimethylation (H3K27Me3). The Nipped-B trimmed mean log2 IP/control values for the individual microarray features are plotted against those for PolII or H3K27Me3. The same 4.25 Mb region of chromosome 2L used for Figs. 2 and 3 is plotted for each comparison, but other regions of the genome show a nearly identical pattern. The top two panels compare Nipped-B to PolII and H3K27Me3 in Sg4 cells, and the bottom panel compares Nipped-B and PolII in BG3 cells. The correlation coefficients (r) for the plotted region are similar to those calculated for the entire non-repetitive genome (Table 1). The plots show that many sequences are enriched by both Nipped-B and PolII immunoprecipitation, but there is less direct correlation in enrichment values at individual features than between Nipped-B and cohesin subunits (Fig. 2), indicating that although Nipped-B and PolII binding overlap, the peaks usually do not align with each other. The middle plot shows that there is essentially no Nipped-B binding to regions with high levels of H3K27Me3
Fig. 7
Binding of Nipped-B and cohesin to the active Abd-B gene in Sg4 cells. The tracks show histone H3 lysine 27 trimethylation (H3K27Me3), PolII, Nipped-B, SA, and Smc1 localization in the bithorax complex (BX-C) for Sg4 cells (black and gray), Smc1 binding in Kc cells (red), and PolII, Nipped-B, and Smc1 binding in BG3 cells (blue). Trimmed mean log2 IP/control values are plotted on a scale from −0.5 to 3. The H3K27Me3 data is from Schwartz et al. 2006. The direction of transcription for the BX-C is indicated with an arrow. An expanded map of Abd-B showing the regulatory region (Akbari et al. 2006; Maeda and Karch 2006) with enhancers (iab, green horizontal lines), boundary elements (Fab, orange vertical bars and arrowheads) and promoter-targeting sequences (PTS, cyan vertical bars) with Nipped-B and PolII binding overlaid on each other illustrates the coincidence of some Nipped-B and PolII peaks. At the lower right is a Northern blot showing Abd-B transcripts in Sg4 cells and their absence in Kc cells. The blot was reprobed for RpL32 as a loading control. Ubx and abd-A transcripts are at least 300-fold lower than Abd-B transcripts in Sg4 cells (Schwartz et al. 2006). Nipped-B and cohesin bind to the Abd-B transcription unit and downstream regulatory region in Sg4 cells where Abd-B is active but not in Kc or BG3 cells where it is silent. The Ubx and abd-A genes, which are actively silenced by Polycomb group proteins in Sg4 cells, as indicated by high H3K27Me3 (Schwartz et al. 2006), do not bind Nipped-B and cohesin in any of the cell lines, although Ubx is regulated by Nipped-B in vivo (Rollins et al. 1999)
Similar articles
- Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells.
Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Gause M, et al. Chromosoma. 2008 Feb;117(1):51-66. doi: 10.1007/s00412-007-0125-5. Epub 2007 Oct 2. Chromosoma. 2008. PMID: 17909832 Free PMC article. - Cohesin, gene expression and development: lessons from Drosophila.
Dorsett D. Dorsett D. Chromosome Res. 2009;17(2):185-200. doi: 10.1007/s10577-009-9022-5. Chromosome Res. 2009. PMID: 19308700 Free PMC article. Review. - Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene.
Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Rollins RA, et al. Mol Cell Biol. 2004 Apr;24(8):3100-11. doi: 10.1128/MCB.24.8.3100-3111.2004. Mol Cell Biol. 2004. PMID: 15060134 Free PMC article. - Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila.
Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Dorsett D, et al. Development. 2005 Nov;132(21):4743-53. doi: 10.1242/dev.02064. Epub 2005 Oct 5. Development. 2005. PMID: 16207752 Free PMC article. - The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
- Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila.
Pherson M, Misulovin Z, Gause M, Dorsett D. Pherson M, et al. Genome Res. 2019 Apr;29(4):602-612. doi: 10.1101/gr.243832.118. Epub 2019 Feb 22. Genome Res. 2019. PMID: 30796039 Free PMC article. - Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole.
Wong RW, Blobel G. Wong RW, et al. Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15441-5. doi: 10.1073/pnas.0807660105. Epub 2008 Oct 1. Proc Natl Acad Sci U S A. 2008. PMID: 18832153 Free PMC article. - The Drosophila enhancer of split gene complex: architecture and coordinate regulation by notch, cohesin, and polycomb group proteins.
Schaaf CA, Misulovin Z, Gause M, Koenig A, Dorsett D. Schaaf CA, et al. G3 (Bethesda). 2013 Oct 3;3(10):1785-94. doi: 10.1534/g3.113.007534. G3 (Bethesda). 2013. PMID: 23979932 Free PMC article. - What fruit flies can tell us about human birth defects.
Dorsett D. Dorsett D. Mo Med. 2013 Jul-Aug;110(4):309-13. Mo Med. 2013. PMID: 24003648 Free PMC article. - Brca2, Pds5 and Wapl differentially control cohesin chromosome association and function.
Misulovin Z, Pherson M, Gause M, Dorsett D. Misulovin Z, et al. PLoS Genet. 2018 Feb 15;14(2):e1007225. doi: 10.1371/journal.pgen.1007225. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29447171 Free PMC article.
References
- Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev Biol. 2006;293:294–304. - PubMed
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol. 2003;13:1941–1953. - PubMed
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM055683-09/GM/NIGMS NIH HHS/United States
- R01 GM055683/GM/NIGMS NIH HHS/United States
- R01 GM070444/GM/NIGMS NIH HHS/United States
- R01GM055683/GM/NIGMS NIH HHS/United States
- P01 HD052860-010003/HD/NICHD NIH HHS/United States
- P01 HD052860/HD/NICHD NIH HHS/United States
- R01GM070444/GM/NIGMS NIH HHS/United States
- P01HD052860/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials