A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity - PubMed (original) (raw)
A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity
Jennifer L Tenor et al. EMBO Rep. 2008 Jan.
Abstract
Pathogen recognition through Toll-like receptors (TLRs) is crucial in order to mount an appropriate immune response against microorganisms. On the basis of a lack of evidence indicating that Caenorhabditis elegans uses TLRs to elicit an immune response and on the absence of genes encoding Rel-like transcription factors in its genome, it is believed that TLR-mediated immunity arose after coelomates split from pseudocoelomates and acoelomates. Here, we show that C. elegans tol-1(nr2033) mutants are killed by the human pathogen Salmonella enterica, which causes a significant pharyngeal invasion in the absence of TOL-1-mediated immunity. We also show that TOL-1 is required for the correct expression of ABF-2, which is a defensin-like molecule expressed in the pharynx, and heat-shock protein 16.41, which is also expressed in the pharynx and is part of a HSP family of proteins required for C. elegans immunity. The results indicate that TOL-1 has a direct role in defence response to certain Gram-negative bacteria and indicate that part of the TLR-mediated immunity might be evolutionarily conserved.
Figures
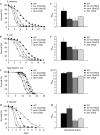
Figure 1
TOL-1-mediated immunity is required for survival in the presence of live Gram-negative bacteria. (A) Wild-type N2 (WT), tol-1(nr2033) (P<0.0001), pmk-1(km25) (P<0.0001) and dbl-1(nk3) (P<0.0001) were exposed to Salmonella enterica. The graphs represent combined results of more than four independent experiments, each of which used 36–50 1-day-old adult hermaphrodites. (B,C) Wild-type N2, tol-1(nr2033), pmk-1(km25) and dbl-1(nk3) were exposed to live (Caenorhabditis elegans killing assay) or heat-killed Escherichia coli (lifespan assay). Although tol-1(nr2033) nematodes died faster than wild type on live E. coli (_P_=0.0024), their lifespan was similar to that of wild type (_P_=0.3966) when exposed to heat-killed E. coli. (D) tol-1(nr2033) lived longer on E. faecalis compared with wild-type (P<0.0001), whereas pmk-1(km25) (P<0.0001) and dbl-1(nk3) (P<0.0001) nematodes were highly susceptible. (E–H) The time for 50% of the nematodes to die (TD50) was determined for each nematode strain exposed to different bacteria. The graphs are representative of at least three independent experiments using 36–50 1-day-old adult hermaphrodites, unless otherwise indicated; bars correspond to mean±s.d.
Figure 2
Salmonella enterica invades the pharynx of tol-1(nr2033). (A–D) Confocal images show the pharynxes of wild-type N2 (WT), pmk-1(km25), dbl-1(nk3) and tol-1(nr2033) nematodes exposed for 48 h to S. enterica expressing green fluorescent protein (GFP). (E,F) Confocal images show the pharynxes of wild-type and tol-1(nr2033) nematodes exposed for 48 h to Pseudomonas aeruginosa expressing GFP. (G) The percentage of nematodes with infected pharynxes was determined for wild type, tol-1(nr2033), pmk-1(km25), dbl-1(nk3) and tol-1(nr2033)Ex[CO7F11;pRF4]; bars correspond to mean±s.d. The Mann–Whitney test indicates that differences among the groups are significantly different; _n_=6–11. (H) Linear regression was used to analyse the relationship between pharyngeal infection and death for wild-type, pmk-1(km25), dbl-1(nk3) and tol-1(nr2033) nematodes exposed to S. enterica expressing GFP. (I) Linear regression was used to analyse the relationship between pharyngeal infection and death for tol-1(nr2033) nematodes exposed to P. aeruginosa or S. enterica expressing GFP. Individual nematodes were monitored daily for pharyngeal infection (24 h before death) and mortality. Images and graphics are representative of 3–11 independent experiments. For each experiment, 20–50 1-day-old adult hermaphrodites were used.
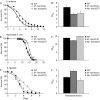
Figure 3
Analysis of the role of candidate downstream components of the TOL-1 pathway in immunity to Salmonella enterica. (A) Wild-type N2 (WT), trf-1(nr2014) (_P_=0.0001) and ikb-1(nr2027) (_P_=0.0001) were exposed to S. enterica. The graphs represent combined results of more than four independent experiments, each of which used 36–50 1-day-old adult hermaphrodites. (B) The lifespan of the ikb-1(nr2027) is similar to that of wild type (_P_=0.3236), whereas the lifespan of trf-1(nr2014) is slightly longer than wild type (_P_=0.0197). (C) trf-1(nr2014) are more resistant to _Enterococcus faecalis-_mediated killing than wild type (P<0.0064), whereas ikb-1(nr2027) (P<0.6730) shows a susceptibility similar to that of wild type. (D–F) The time for 50% of the nematodes to die (TD50) was determined for each nematode strain exposed to different bacteria. The graphs are representative of at least three independent experiments, each of which used 36–50 1-day-old adult hermaphrodites; bars correspond to mean±s.d., unless otherwise indicated.
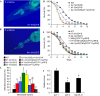
Figure 4
Characterization of downstream components of the TOL-1 pathway. (A,B) Confocal images show the pharynxes of trf-1(nr2014) and ikb-1(nr2027) nematodes exposed for 48 h to Salmonella enterica expressing green fluorescent protein (GFP). GFP-expressing bacteria were detected in the pharynx and intestinal lumen of both mutants. (C) The percentage of nematodes with infected pharynxes was determined for wild type (WT), trf-1(nr2014), ikb-1(nr2027), tol-1(nr2033), tol-1(nr2033); trf-1(nr2014), trf-1(nr2014) Ex[CO7F11;pRF4] and ikb-1(nr2027)Ex[CO7F11;pRF4]. For each experiment, 20–50 1-day-old adult hermaphrodites were used. The Mann–Whitney test indicates that differences among the groups are significantly different; n_=3–11. (D,E) Wild-type N2, tol-1(nr2033) (P<0.0001), trf-1(nr2014) (P_=0.0013), tol-1(nr2033);trf-1(nr2014) (P<0.0001), wild-type Ex[CO7F11;pRF4] (P<0.0024), trf-1(nr2014)Ex[CO7F11;pRF4] (P<0.0015) and ikb-1(nr2027)Ex[CO7F11;pRF4] (P<0.002) were exposed to S. enterica. The graphs represent combined results of more than four independent experiments, each of which used 36–50 1-day-old adult hermaphrodites. (F) Quantitative reverse transcription–PCR analysis of act-1, abf-2 and hsp-16.41 expression in tol-1(nr2033) relative to wild-type nematodes grown on S. enterica. Data were analysed by relative quantitation using the comparative cycle threshold method and normalization to act-1. Student's exact _t_-test indicates that differences among the groups are significantly different; _n_=3; bars correspond to mean±s.d.
Similar articles
- Origin of Toll-like receptor-mediated innate immunity.
Kanzok SM, Hoa NT, Bonizzoni M, Luna C, Huang Y, Malacrida AR, Zheng L. Kanzok SM, et al. J Mol Evol. 2004 Apr;58(4):442-8. doi: 10.1007/s00239-003-2565-8. J Mol Evol. 2004. PMID: 15114422 - A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans.
Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. Pujol N, et al. Curr Biol. 2001 Jun 5;11(11):809-21. doi: 10.1016/s0960-9822(01)00241-x. Curr Biol. 2001. PMID: 11516642 - Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway.
Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Aballay A, et al. Curr Biol. 2003 Jan 8;13(1):47-52. doi: 10.1016/s0960-9822(02)01396-9. Curr Biol. 2003. PMID: 12526744 - Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates.
Irazoqui JE, Urbach JM, Ausubel FM. Irazoqui JE, et al. Nat Rev Immunol. 2010 Jan;10(1):47-58. doi: 10.1038/nri2689. Nat Rev Immunol. 2010. PMID: 20029447 Free PMC article. Review. - Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans.
Kim DH, Ausubel FM. Kim DH, et al. Curr Opin Immunol. 2005 Feb;17(1):4-10. doi: 10.1016/j.coi.2004.11.007. Curr Opin Immunol. 2005. PMID: 15653303 Review.
Cited by
- Dissociation of immune responses from pathogen colonization supports pattern recognition in C. elegans.
Twumasi-Boateng K, Shapira M. Twumasi-Boateng K, et al. PLoS One. 2012;7(4):e35400. doi: 10.1371/journal.pone.0035400. Epub 2012 Apr 13. PLoS One. 2012. PMID: 22514739 Free PMC article. - C. elegans orphan nuclear receptor NHR-42 represses innate immunity and promotes lipid loss downstream of HLH-30/TFEB.
Goswamy D, Gonzalez X, Labed SA, Irazoqui JE. Goswamy D, et al. Front Immunol. 2023 Feb 13;14:1094145. doi: 10.3389/fimmu.2023.1094145. eCollection 2023. Front Immunol. 2023. PMID: 36860863 Free PMC article. - Caenorhabditis elegans: a model to understand host-microbe interactions.
Kumar A, Baruah A, Tomioka M, Iino Y, Kalita MC, Khan M. Kumar A, et al. Cell Mol Life Sci. 2020 Apr;77(7):1229-1249. doi: 10.1007/s00018-019-03319-7. Epub 2019 Oct 4. Cell Mol Life Sci. 2020. PMID: 31584128 Free PMC article. Review. - Caenorhabditis elegans in high-throughput screens for anti-infective compounds.
Peterson ND, Pukkila-Worley R. Peterson ND, et al. Curr Opin Immunol. 2018 Oct;54:59-65. doi: 10.1016/j.coi.2018.06.003. Epub 2018 Jun 20. Curr Opin Immunol. 2018. PMID: 29935375 Free PMC article. Review. - Dopamine modulates acetylcholine release via octopamine and CREB signaling in Caenorhabditis elegans.
Suo S, Ishiura S. Suo S, et al. PLoS One. 2013 Aug 16;8(8):e72578. doi: 10.1371/journal.pone.0072578. eCollection 2013. PLoS One. 2013. PMID: 23977320 Free PMC article.
References
- Aballay A, Yorgey P, Ausubel FM (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol 10: 1539–1542 - PubMed
- Aballay A, Drenkard E, Hilbun LR, Ausubel FM (2003) Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol 13: 47–52 - PubMed
- Apfeld J, Kenyon C (1998) Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95: 199–210 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM070977/GM/NIGMS NIH HHS/United States
- R21 AI065641/AI/NIAID NIH HHS/United States
- GM 070977/GM/NIGMS NIH HHS/United States
- R37 GM070977/GM/NIGMS NIH HHS/United States
- U54 AI 057157/AI/NIAID NIH HHS/United States
- AI 065641/AI/NIAID NIH HHS/United States
- U54 AI057157/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases