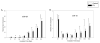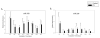Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis - PubMed (original) (raw)
Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis
Hana Bruchova et al. Exp Hematol. 2007 Nov.
Abstract
Objective: Polycythemia vera (PV) is a myeloproliferative disorder, arising from the acquired mutation(s) of a hematopoietic stem cell. The JAK2 V617F somatic mutation is found in most PV patients; however, it is not the disease-initiating mutation. Because microRNAs (miRNAs) play a regulatory role in hematopoiesis, we studied miRNA expressions in PV and normal erythropoiesis.
Methods: Peripheral blood mononuclear cells were cultured in a three-phase liquid system resulting in synchronized expansion of erythroid progenitors. Using gene-expression profiling by CombiMatrix MicroRNArray, we searched for PV-specific changes at days 1, 14, and 21. Twelve miRNA candidates were then reevaluated by quantitative real-time polymerase chain reaction in a larger number of samples obtained from progenitors at the same stage of differentiation.
Results: A significant difference in miR-150 expression was found in PV. In normal erythropoiesis, three expression patterns of miRNAs were observed: progressive downregulation of miR-150, miR-155, miR-221, miR-222; upregulation of miR-451, miR-16 at late stages of erythropoiesis; and biphasic regulation of miR-339, miR-378. The miR-451 appears to be erythroid-specific.
Conclusions: We identified the miRNAs with regulated expression in erythropoiesis; one appeared to be PV-specific. Their miRNA expression levels define early, intermediate, and late stages of erythroid differentiation. The validity of our findings was confirmed in nonexpanded peripheral blood cells.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
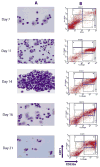
Figure 1. Induction of erythroid differentiation from peripheral blood MNCs
A) Morphology analysis of Wright-Giemsa stained cells at selected days (magnification 100×). The representative populations are shown: day7 – immature early erythroid progenitors, day11 - proerythroblasts and basophilic erythroblasts, day14 - mostly basophilic erythroblasts, day16 - basophilic and polychromatophilic erythroblasts, day21 – late normoblasts and some reticulocytes; B) Flow cytometry analysis of surface antigens, CD71 and CD235a, on expanded cells at different stages of differentiation. The cells were immunostained with (FITC)-conjugated anti-CD71 and (PE)-conjugated anti-CD235a antibodies. In the plot, X- and Y-axes indicate the relative fluorescence of PE and FITC, respectively. The gated regions define characteristic expression patterns of the surface antigens: CD71med and CD235alow (R1), - CD71high and CD235alow (R2), - CD71high and CD235ahigh (R3), - CD71med and CD235ahigh (R4), - CD71low and CD235ahigh (R5).
Figure 2. Expression of the differentially regulated miRNAs during erythroid differentiation
The one-way ANOVA analysis (P<0.01) selected 30 miRNAs which were differentially expressed between the 3 groups (Day1 versus Day14 versus Day21). The control samples from day21 were pooled because of limited amount of RNA. The relative gene expressions are expressed by a gradient intensity of color, as shown in the grey scale at the top. The darkest color indicates no expression and the lightest color indicates maximal expression. C-control, PV- polycythemia vera patient.
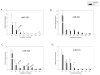
Figure 3. Gene expression patterns of the miRNAs depicting increased levels during erythroid differentiation
Gene expression was determined by qRT-PCR at the defined time points. Relative gene expression was calculated based on generation of standard curve and normalized against the endogenous control RNAU6B. The plotted data (arbitrary units) express as mean with standard error. Statistical significance between control and PV cells was calculated by t-test at each time point (*P<0.05, **P<0.01). Control - healthy controls, PV- polycythemia vera patients, D – day.
Figure 4. Gene expression patterns of the miRNAs depicting decreased levels during erythroid differentiation
Gene expression was determined by qRT-PCR at the defined time points. Relative gene expression was calculated based on generation of standard curve and normalized against the endogenous control RNAU6B. The plotted data (arbitrary units) express as mean with standard error. Statistical significance between control and PV cells was calculated by t-test at each time point (*P<0.05, **P<0.01). Control - healthy controls, PV- polycythemia vera patients, D – day.
Figure 5. Gene expression patterns of the miRNAs depicting biphasic regulation during erythroid differentiation
Gene expression was determined by qRT-PCR at the defined time points. Relative gene expression was calculated based on generation of standard curve and normalized against the endogenous control RNAU6B. The plotted data (arbitrary units) express as mean with standard error. Statistical significance between control and PV cells was calculated by t-test at each time point (*P<0.05, **P<0.01). Control - healthy controls, PV- polycythemia vera patients, D – day.
Figure 6. Expression levels of the miRNAs in non-expanded peripheral blood cells from controls and PV patients
Gene expression was determined by qRT-PCR. Relative gene expression was calculated based on generation of standard curve and normalized against the endogenous control RNAU6B. The plotted data (arbitrary units) express as mean with standard error. Statistical significance between control and PV cells was calculated by t-test in each cell lineage (*P<0.05, **P<0.01). Control - healthy controls, PV - polycythemia vera patients, MNC - mononuclear cells, GRAN - granulocytes, RETIC - reticulocytes, PLAT - platelets.
Figure 6. Expression levels of the miRNAs in non-expanded peripheral blood cells from controls and PV patients
Gene expression was determined by qRT-PCR. Relative gene expression was calculated based on generation of standard curve and normalized against the endogenous control RNAU6B. The plotted data (arbitrary units) express as mean with standard error. Statistical significance between control and PV cells was calculated by t-test in each cell lineage (*P<0.05, **P<0.01). Control - healthy controls, PV - polycythemia vera patients, MNC - mononuclear cells, GRAN - granulocytes, RETIC - reticulocytes, PLAT - platelets.
Similar articles
- Erythropoiesis in polycythemia vera is hyper-proliferative and has accelerated maturation.
Bruchova H, Yoon D, Agarwal AM, Swierczek S, Prchal JT. Bruchova H, et al. Blood Cells Mol Dis. 2009 Jul-Aug;43(1):81-7. doi: 10.1016/j.bcmd.2009.02.001. Epub 2009 Mar 4. Blood Cells Mol Dis. 2009. PMID: 19264517 Free PMC article. - Aberrant expression of microRNA in polycythemia vera.
Bruchova H, Merkerova M, Prchal JT. Bruchova H, et al. Haematologica. 2008 Jul;93(7):1009-16. doi: 10.3324/haematol.12706. Epub 2008 May 27. Haematologica. 2008. PMID: 18508790 - MicroRNA deregulation in polycythemia vera and essential thrombocythemia patients.
Zhan H, Cardozo C, Yu W, Wang A, Moliterno AR, Dang CV, Spivak JL. Zhan H, et al. Blood Cells Mol Dis. 2013 Mar;50(3):190-5. doi: 10.1016/j.bcmd.2012.11.009. Epub 2012 Dec 21. Blood Cells Mol Dis. 2013. PMID: 23265742 Free PMC article. - Polycythemia vera: scientific advances and current practice.
Tefferi A, Spivak JL. Tefferi A, et al. Semin Hematol. 2005 Oct;42(4):206-20. doi: 10.1053/j.seminhematol.2005.08.003. Semin Hematol. 2005. PMID: 16210034 Review.
Cited by
- The Role of MicroRNAs in Hematopoietic Stem Cell and Leukemic Stem Cell Function.
Chung SS, Hu W, Park CY. Chung SS, et al. Ther Adv Hematol. 2011 Oct;2(5):317-34. doi: 10.1177/2040620711410772. Ther Adv Hematol. 2011. PMID: 23556099 Free PMC article. - A Novel Role for Progesterone Receptor Membrane Component 1 (PGRMC1): A Partner and Regulator of Ferrochelatase.
Piel RB 3rd, Shiferaw MT, Vashisht AA, Marcero JR, Praissman JL, Phillips JD, Wohlschlegel JA, Medlock AE. Piel RB 3rd, et al. Biochemistry. 2016 Sep 20;55(37):5204-17. doi: 10.1021/acs.biochem.6b00756. Epub 2016 Sep 9. Biochemistry. 2016. PMID: 27599036 Free PMC article. - Implication of microRNAs in the pathogenesis of MDS.
Fang J, Varney M, Starczynowski DT. Fang J, et al. Curr Pharm Des. 2012;18(22):3170-9. doi: 10.2174/1381612811209023170. Curr Pharm Des. 2012. PMID: 22571695 Free PMC article. Review. - Serum miR-222-3p as a Double-Edged Sword in Predicting Efficacy and Trastuzumab-Induced Cardiotoxicity for HER2-Positive Breast Cancer Patients Receiving Neoadjuvant Target Therapy.
Zhang S, Wang Y, Wang Y, Peng J, Yuan C, Zhou L, Xu S, Lin Y, Du Y, Yang F, Zhang J, Dai H, Yin W, Lu J. Zhang S, et al. Front Oncol. 2020 Apr 28;10:631. doi: 10.3389/fonc.2020.00631. eCollection 2020. Front Oncol. 2020. PMID: 32426280 Free PMC article. - Upregulation of miR‑6747‑3p affects red blood cell lineage development and induces fetal hemoglobin expression by targeting BCL11A in β‑thalassemia.
Lv A, Chen M, Zhang S, Zhao W, Li J, Lin S, Zheng Y, Lin N, Xu L, Huang H. Lv A, et al. Mol Med Rep. 2025 Jan;31(1):7. doi: 10.3892/mmr.2024.13372. Epub 2024 Oct 25. Mol Med Rep. 2025. PMID: 39450557 Free PMC article.
References
- Bellucci S, Michiels JJ. The role of JAK2 V617F mutation, spontaneous erythropoiesis and megakaryocytopoiesis, hypersensitive platelets, activated leukocytes, and endothelial cells in the etiology of thrombotic manifestations in polycythemia vera and essential thrombocythemia. Semin Thromb Hemost. 2006;32:381–398. - PubMed
- Levine RL, Gilliland DG. JAK-2 mutations and their relevance to myeloproliferative disease. Curr Opin Hematol. 2007;14:43–47. - PubMed
- Rumi E, Passamonti F, Pietra D, et al. JAK2 (V617F) as an acquired somatic mutation and a secondary genetic event associated with disease progression in familial myeloproliferative disorders. Cancer. 2006;107:2206–2211. - PubMed
- Lippert E, Boissinot M, Kralovics R, et al. The JAK2-V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood. 2006;108:1865–1867. - PubMed
- Nussenzveig RH, Swierczek SI, Jelinek J, et al. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol. 2007;35:32–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA108671/CA/NCI NIH HHS/United States
- R01 HL050077/HL/NHLBI NIH HHS/United States
- NR/9236-3/NR/NINR NIH HHS/United States
- 1P01CA108671-O1A2/CA/NCI NIH HHS/United States
- R01HL50077-11/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous