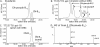One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica - PubMed (original) (raw)
One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica
Peter J Anderson et al. J Bacteriol. 2008 Feb.
Abstract
Corrinoid (vitamin B12-like) cofactors contain various alpha-axial ligands, including 5,6-dimethylbenzimidazole (DMB) or adenine. The bacterium Salmonella enterica produces the corrin ring only under anaerobic conditions, but it can form "complete" corrinoids aerobically by importing an "incomplete" corrinoid, such as cobinamide (Cbi), and adding appropriate alpha- and beta-axial ligands. Under aerobic conditions, S. enterica performs the corrinoid-dependent degradation of ethanolamine if given vitamin B12, but it can make B12 from exogenous Cbi only if DMB is also provided. Mutants isolated for their ability to degrade ethanolamine without added DMB converted Cbi to pseudo-B12 cofactors (having adenine as an alpha-axial ligand). The mutations cause an increase in the level of free adenine and install adenine (instead of DMB) as an alpha-ligand. When DMB is provided to these mutants, synthesis of pseudo-B12 cofactors ceases and B12 cofactors are produced, suggesting that DMB regulates production or incorporation of free adenine as an alpha-ligand. Wild-type cells make pseudo-B12 cofactors during aerobic growth on propanediol plus Cbi and can use pseudo-vitamin B12 for all of their corrinoid-dependent enzymes. Synthesis of coenzyme pseudo-B12 cofactors requires the same enzymes (CobT, CobU, CobS, and CobC) that install DMB in the formation of coenzyme B12. Models are described for the mechanism and control of alpha-axial ligand installation.
Figures
FIG. 1.
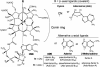
Alternative “complete” corrinoids in S. enterica. The CN(β) derivatives are formed in the process of extracting corrinoids from cells. A methyl group (not shown) can also serve as a β-axial ligand and is involved in methyl transfer reactions. The corrin moiety is provided to cells as dicyanocobinamide (CN)2Cbi, which has only the aminopropanol side chain and no ligand nucleotide; in the presence of CN− ions, it has CN as both the β- and α-axial ligand.
FIG. 2.
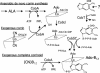
Pathways for formation of complete corrinoids. In the anaerobic de novo pathway, the β-axial ligand adenosine is added to cobyrinic acid diamide (CobA), followed by four amidations and addition of the aminopropanol side chain to produce Ado-Cbi. Under aerobic conditions, the corrin ring is supplied as (CN)2Cbi, which is adenosylated by CobA to produce Ado-Cbi. The aminopropanol side chain of this corrin is activated by the addition of GDP (CobU), and the α-axial ligand loop is completed by replacing GMP with a nucleoside of either DMB or adenine (CobS and CobC), thereby generating a complete corrinoid. CobT produces the dinucleotide that donates the nucleoside group. Note that the corrin ring compound (CN)2Cbi is not the natural substrate for the CobA enzyme. Ade, adenine; ALA, aminolevulinic acid; Co, corrin; Gua, guanine; Nm, nicotinamide.
FIG. 3.
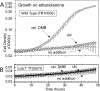
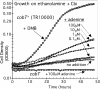
DMB-dependent aerobic growth on ethanolamine. (A) Aerobic growth of wild-type S. enterica (TR10000) on minimal NCE medium with additions as shown. (B) A cobT mutant (TT25575) cannot grow even when both Cbi and DMB are provided. Culture OD600 was monitored using a BioTek Synergy HT plate reader and KC4 software; it is presented on a logarithmic scale.
FIG. 4.
Growth of DMB-independent apt and amn mutants. Growth was tested aerobically on minimal NCE medium with ethanolamine as the sole carbon source, and growth was monitored at OD600. (A) Strain TT25778 carries the constructed deletion mutation apt-18. (B) Strain TT25779 carries the apt-18 deletion and a cobT insertion mutation. (C) Strain TT25112 carries the amnA52 point mutation (G935A). (D) Strain TT25777 carries both the amnA52 point mutation and a cobT insertion.
FIG. 5.
HPLC/MS of cyanidated extracts from the apt-18 deletion mutant (TT25778). (A) Separation of standards. CN-pseudo-B12 elutes at 19.39 min, and vitamin B12 (CN-Cbl) elutes at 20.03 min under the chromatographic conditions used for all separations. (B) Cyanidated extracts from cells grown on ethanolamine and cobinamide. Peak 1 (16.97 min), α-CN-pseudo-B12 isomer; peak 2 (17.60 min), CN-cobinamide-GDP isomer; peak 3 (19.38 min), CN-pseudo-B12; peak 4 (20.03 min), CN-cobinamide phosphate; peak 5 (21.07 min), CN-cobinamide-GDP. (C) Cyanidated extracts from cells grown on ethanolamine and cobinamide in the presence of DMB. The peaks are vitamin B12 (20.00 min) and B12 isomer (21.55 min). AU, absorbance units. (D) Electrospray ionization mass spectrum of peak 3 having a molecular ion (M+H)+ of 1344.5453, the exact mass of CN-pseudo-B12 (change of 0.5 ppm). Peaks with standard β-CN-corrinoid forms are indicated by solid black arrow; alternative α-CN isomers are indicated with dashed lines.
FIG. 6.
Effect of adenine base on the ability to use ethanolamine. Wild-type cells were tested for the ability to grow aerobically on minimal NCE medium with ethanolamine as the sole carbon source and the indicated supplementation by adenine. Culture OD600 was monitored using a BioTek Synergy HT plate reader and KC4 software and is presented on a logarithmic scale.
FIG. 7.
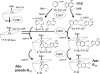
Proposed pathway for completion of Ado-pseudo-B12 and Ado-Cbl. In this pathway, the reaction at the left forming AdoCbi-P-P-5R1-Gua is well established. The lower ligand base (adenine or DMB) is exchanged for the nicotinamide (Nm) moiety of NAD to form the donor dinucleotide (either α-7AAD or α-DAD). It is proposed that CobS catalyzes an attack on the pyrophosphate of the activated corrin precursor by the 3′OH of the ribose nearest the critical base (adenine or DMB) of the dinucleotide. This forms the proposed penultimate intermediate whose ribose has substitutions at positions 1, 3, and 5. Removal of AMP (remainder of the donor dinucleotide) from the 5′ position leaves the complete corrinoid. Ade, adenine; Co, corrinoid; Gua, guanine.
Comment in
- Pseudo-B12 joins the cofactor family.
Taga ME, Walker GC. Taga ME, et al. J Bacteriol. 2008 Feb;190(4):1157-9. doi: 10.1128/JB.01892-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083805 Free PMC article. No abstract available.
Similar articles
- Cobalamin production by Lactobacillus coryniformis: biochemical identification of the synthetized corrinoid and genomic analysis of the biosynthetic cluster.
Torres AC, Vannini V, Bonacina J, Font G, Saavedra L, Taranto MP. Torres AC, et al. BMC Microbiol. 2016 Oct 13;16(1):240. doi: 10.1186/s12866-016-0854-9. BMC Microbiol. 2016. PMID: 27737643 Free PMC article. - Pseudo-B12 joins the cofactor family.
Taga ME, Walker GC. Taga ME, et al. J Bacteriol. 2008 Feb;190(4):1157-9. doi: 10.1128/JB.01892-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083805 Free PMC article. No abstract available. - The end of the cob operon: evidence that the last gene (cobT) catalyzes synthesis of the lower ligand of vitamin B12, dimethylbenzimidazole.
Chen P, Ailion M, Weyand N, Roth J. Chen P, et al. J Bacteriol. 1995 Mar;177(6):1461-9. doi: 10.1128/jb.177.6.1461-1469.1995. J Bacteriol. 1995. PMID: 7883701 Free PMC article. - Biochemistry of B12-cofactors in human metabolism.
Kräutler B. Kräutler B. Subcell Biochem. 2012;56:323-46. doi: 10.1007/978-94-007-2199-9_17. Subcell Biochem. 2012. PMID: 22116707 Review. - Methylations in vitamin B12 biosynthesis and catalysis.
Mathur Y, Hazra AB. Mathur Y, et al. Curr Opin Struct Biol. 2022 Dec;77:102490. doi: 10.1016/j.sbi.2022.102490. Epub 2022 Nov 10. Curr Opin Struct Biol. 2022. PMID: 36371846 Review.
Cited by
- Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi.
Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. Yi S, et al. Appl Environ Microbiol. 2012 Nov;78(21):7745-52. doi: 10.1128/AEM.02150-12. Epub 2012 Aug 24. Appl Environ Microbiol. 2012. PMID: 22923412 Free PMC article. - A novel regulatory interplay between atypical B12 riboswitches and uORF translation in Mycobacterium tuberculosis.
Kipkorir T, Polgar P, Barker D, D'Halluin A, Patel Z, Arnvig KB. Kipkorir T, et al. Nucleic Acids Res. 2024 Jul 22;52(13):7876-7892. doi: 10.1093/nar/gkae338. Nucleic Acids Res. 2024. PMID: 38709884 Free PMC article. - De Novo Cobalamin Biosynthesis, Transport, and Assimilation and Cobalamin-Mediated Regulation of Methionine Biosynthesis in Mycobacterium smegmatis.
Kipkorir T, Mashabela GT, de Wet TJ, Koch A, Dawes SS, Wiesner L, Mizrahi V, Warner DF. Kipkorir T, et al. J Bacteriol. 2021 Mar 8;203(7):e00620-20. doi: 10.1128/JB.00620-20. Print 2021 Mar 8. J Bacteriol. 2021. PMID: 33468593 Free PMC article. - The Distribution of SIgA and IgG Antibody-Secreting Cells in the Small Intestine of Bactrian Camels (Camelus bactrianus) of Different Ages.
Zhang WD, Wang WH, Jia S. Zhang WD, et al. PLoS One. 2016 Jun 1;11(6):e0156635. doi: 10.1371/journal.pone.0156635. eCollection 2016. PLoS One. 2016. PMID: 27249417 Free PMC article. - Bacillus megaterium has both a functional BluB protein required for DMB synthesis and a related flavoprotein that forms a stable radical species.
Collins HF, Biedendieck R, Leech HK, Gray M, Escalante-Semerena JC, McLean KJ, Munro AW, Rigby SE, Warren MJ, Lawrence AD. Collins HF, et al. PLoS One. 2013;8(2):e55708. doi: 10.1371/journal.pone.0055708. Epub 2013 Feb 14. PLoS One. 2013. PMID: 23457476 Free PMC article.
References
- Carkeet, C., S. R. Dueker, J. Lango, B. A. Buchholz, J. W. Miller, R. Green, B. D. Hammock, J. R. Roth, and P. J. Anderson. 2006. Human vitamin B12 absorption measurement by accelerator mass spectrometry using specifically labeled 14C-cobalamin. Proc. Natl. Acad. Sci. USA 1035694-5699. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5 P42 ES04699/ES/NIEHS NIH HHS/United States
- P42 ES004699/ES/NIEHS NIH HHS/United States
- GM34804/GM/NIGMS NIH HHS/United States
- P01 ES11269/ES/NIEHS NIH HHS/United States
- P01 ES011269/ES/NIEHS NIH HHS/United States
- R01 GM034804/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases