DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae) - PubMed (original) (raw)
DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae)
Mark A Chapman et al. BMC Plant Biol. 2007.
Abstract
Background: Safflower (Carthamus tinctorius L.) is a diploid oilseed crop whose origin is largely unknown. Safflower is widely believed to have been domesticated over 4,000 years ago somewhere in the Fertile Crescent. Previous hypotheses regarding the origin of safflower have focused primarily on two other species from sect. Carthamus - C. oxyacanthus and C. palaestinus - as the most likely progenitors, although some attention has been paid to a third species (C. persicus) as a possible candidate. Here, we describe the results of a phylogenetic analysis of the entire section using data from seven nuclear genes.
Results: Single gene phylogenetic analyses indicated some reticulation or incomplete lineage sorting. However, the analysis of the combined dataset revealed a close relationship between safflower and C. palaestinus. In contrast, C. oxyacanthus and C. persicus appear to be more distantly related to safflower.
Conclusion: Based on our results, we conclude that safflower is most likely derived from the wild species Carthamus palaestinus. As expected, safflower exhibits somewhat reduced nucleotide diversity as compared to its progenitor, consistent with the occurrence of a population genetic bottleneck during domestication. The results of this research set the stage for an investigation of the genetics of safflower domestication.
Figures
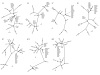
Figure 1
Phylogenetic relationships among species of Carthamus sect. Carthamus based on single-gene analyses. Neighbor-Joining trees were generated for each individual gene. Species names and accession codes are given in Table 1. Accession names followed by -1 or -2 denote alleles for a given locus. Alleles followed by a * were determined using haplotype subtraction by maximum likelihood; the remainder of the alleles were determined by cloning.
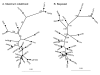
Figure 2
Phylogenetic relationships among species of Carthamus sect. Carthamus based on a combined analysis of seven nuclear genes. Maximum likelihood (A) and Bayesian (B) trees generated for the combined dataset. Bootstrap values (> 75%) for the ML tree and posterior probabilities (> 0.90) for the Bayesian tree are given alongside branches. Species names and accession codes are given in Table 1.
Similar articles
- Phylogenomic investigation of safflower (Carthamus tinctorius) and related species using genotyping-by-sequencing (GBS).
Sardouei-Nasab S, Nemati Z, Mohammadi-Nejad G, Haghi R, Blattner FR. Sardouei-Nasab S, et al. Sci Rep. 2023 Apr 17;13(1):6212. doi: 10.1038/s41598-023-33347-0. Sci Rep. 2023. PMID: 37069212 Free PMC article. - Comprehensive review of two groups of flavonoids in Carthamus tinctorius L.
Xian B, Wang R, Jiang H, Zhou Y, Yan J, Huang X, Chen J, Wu Q, Chen C, Xi Z, Ren C, Pei J. Xian B, et al. Biomed Pharmacother. 2022 Sep;153:113462. doi: 10.1016/j.biopha.2022.113462. Epub 2022 Aug 1. Biomed Pharmacother. 2022. PMID: 36076573 Review. - Extraction, Structures, Bioactivities and Structure-Function Analysis of the Polysaccharides From Safflower (Carthamus tinctorius L.).
Wu X, Cai X, Ai J, Zhang C, Liu N, Gao W. Wu X, et al. Front Pharmacol. 2021 Oct 20;12:767947. doi: 10.3389/fphar.2021.767947. eCollection 2021. Front Pharmacol. 2021. PMID: 34744747 Free PMC article. Review.
Cited by
- Identification of genes associated with fatty acid biosynthesis based on 214 safflower core germplasm.
Fan K, Qin Y, Hu X, Xu J, Ye Q, Zhang C, Ding Y, Li G, Chen Y, Liu J, Wang P, Hu Z, Yan X, Xiong H, Liu H, Qin R. Fan K, et al. BMC Genomics. 2023 Dec 11;24(1):763. doi: 10.1186/s12864-023-09874-5. BMC Genomics. 2023. PMID: 38082219 Free PMC article. - Analysis of the first Taraxacum kok-saghyz transcriptome reveals potential rubber yield related SNPs.
Luo Z, Iaffaldano BJ, Zhuang X, Fresnedo-Ramírez J, Cornish K. Luo Z, et al. Sci Rep. 2017 Aug 30;7(1):9939. doi: 10.1038/s41598-017-09034-2. Sci Rep. 2017. PMID: 28855528 Free PMC article. - The chromosome-scale reference genome of safflower (Carthamus tinctorius) provides insights into linoleic acid and flavonoid biosynthesis.
Wu Z, Liu H, Zhan W, Yu Z, Qin E, Liu S, Yang T, Xiang N, Kudrna D, Chen Y, Lee S, Li G, Wing RA, Liu J, Xiong H, Xia C, Xing Y, Zhang J, Qin R. Wu Z, et al. Plant Biotechnol J. 2021 Sep;19(9):1725-1742. doi: 10.1111/pbi.13586. Epub 2021 Apr 8. Plant Biotechnol J. 2021. PMID: 33768699 Free PMC article. - Introgression potential between safflower (Carthamus tinctorius) and wild relatives of the genus Carthamus.
Mayerhofer M, Mayerhofer R, Topinka D, Christianson J, Good AG. Mayerhofer M, et al. BMC Plant Biol. 2011 Mar 14;11:47. doi: 10.1186/1471-2229-11-47. BMC Plant Biol. 2011. PMID: 21401959 Free PMC article. - The use of a candidate gene approach to study Botrytis cinerea resistance in Gerbera hybrida.
Fu Y, Song Y, van Tuyl JM, Visser RGF, Arens P. Fu Y, et al. Front Plant Sci. 2023 Mar 22;14:1100416. doi: 10.3389/fpls.2023.1100416. eCollection 2023. Front Plant Sci. 2023. PMID: 37035068 Free PMC article.
References
- Knowles PF, Ashri A. Safflower: Carthamus tinctorius (Compositae) In: Smartt J, Simmonds NW, editor. Evolution of Crop Plants. 2nd. Harlow, UK , Longman; 1995. pp. 47–50.
- Smith JR. Safflower. Champaign, IL. , AOCS Press; 1996.
- Knowles PF. Centers of plant diversity and conservation of crop germplasm - Safflower. Economic Botany. 1969;23:324–329.
- Weiss EA. Castor, Sesame and Safflower. New York, NY , Barnes and Noble, Inc.; 1971.
- Knowles PF. Safflower. Advances in Agronomy. 1958;10:289–323.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials