Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus - PubMed (original) (raw)
Comparative Study
doi: 10.1186/1471-2180-7-99.
Kristina G Hultén, Xiang Qin, Huaiyang Jiang, Shailaja Yerrapragada, Edward O Mason Jr, Yue Shang, Tiffany M Williams, Régine M Fortunov, Yamei Liu, Okezie Igboeli, Joseph Petrosino, Madhan Tirumalai, Akif Uzman, George E Fox, Ana Maria Cardenas, Donna M Muzny, Lisa Hemphill, Yan Ding, Shannon Dugan, Peter R Blyth, Christian J Buhay, Huyen H Dinh, Alicia C Hawes, Michael Holder, Christie L Kovar, Sandra L Lee, Wen Liu, Lynne V Nazareth, Qiaoyan Wang, Jianling Zhou, Sheldon L Kaplan, George M Weinstock
Affiliations
- PMID: 17986343
- PMCID: PMC2222628
- DOI: 10.1186/1471-2180-7-99
Comparative Study
Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus
Sarah K Highlander et al. BMC Microbiol. 2007.
Abstract
Background: Community acquired (CA) methicillin-resistant Staphylococcus aureus (MRSA) increasingly causes disease worldwide. USA300 has emerged as the predominant clone causing superficial and invasive infections in children and adults in the USA. Epidemiological studies suggest that USA300 is more virulent than other CA-MRSA. The genetic determinants that render virulence and dominance to USA300 remain unclear.
Results: We sequenced the genomes of two pediatric USA300 isolates: one CA-MRSA and one CA-methicillin susceptible (MSSA), isolated at Texas Children's Hospital in Houston. DNA sequencing was performed by Sanger dideoxy whole genome shotgun (WGS) and 454 Life Sciences pyrosequencing strategies. The sequence of the USA300 MRSA strain was rigorously annotated. In USA300-MRSA 2658 chromosomal open reading frames were predicted and 3.1 and 27 kilobase (kb) plasmids were identified. USA300-MSSA contained a 20 kb plasmid with some homology to the 27 kb plasmid found in USA300-MRSA. Two regions found in US300-MRSA were absent in USA300-MSSA. One of these carried the arginine deiminase operon that appears to have been acquired from S. epidermidis. The USA300 sequence was aligned with other sequenced S. aureus genomes and regions unique to USA300 MRSA were identified.
Conclusion: USA300-MRSA is highly similar to other MRSA strains based on whole genome alignments and gene content, indicating that the differences in pathogenesis are due to subtle changes rather than to large-scale acquisition of virulence factor genes. The USA300 Houston isolate differs from another sequenced USA300 strain isolate, derived from a patient in San Francisco, in plasmid content and a number of sequence polymorphisms. Such differences will provide new insights into the evolution of pathogens.
Figures
Figure 1
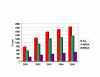
Histogram of the prevalence of community-acquired MR and MS strains of S. aureus at TCH over a four-year study. All cases are shown in red, MR cases in green and MS cases in purple.
Figure 2
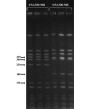
PFGE of Sma I-digested USA300-HOU-MR and USA300-HOU-MS. Replicates of MR (left) and MS (right) profiles are shown. Molecular weight markers are shown in kilobases.
Figure 3
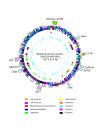
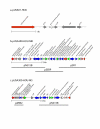
Chromosomal map of USA300-HOU-MR with landmarks indicating the SCC_mec_-ACME region (outer circle, green), pathogenicity islands (outer circle, light blue), prophage (outer circle, lavender), the 13 kb insertion sequence (outer circle, orange), and two regions of sequence not found in USA300-HOU-MS (outer circle, yellow). ORFs on both strands are represented by the second circle and are colored according to functional categories as follows and as shown in the key: cell processes, tan; cell structure, violet; DNA replication and recombination, red; general metabolism, blue; regulation, green; transcription, yellow; translation, orange; transport, cyan; virulence, fuchsia; unknown, black. The third circle shows RNAs: rRNAs, blue; tRNAs, red; ncRNAs, green. The inner circle shows the location of SNPs between USA300-HOU-MR and FPR3757.
Figure 4
Linear representation of BLASTN comparison of S. aureus strains to USA300-HOU-MR. Strains are listed on the y-axis and the x-axis shows the USA300-HOU-MR coordinates. Bars represent regions of at least 50 bp in length present in USA300-HOU-MR but absent in the other strains.
Figure 5
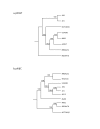
Boot-strapped phylogenetic trees of the prophages found in USA300-HOU-MR: a) phiSLT phage and b) phiβC family phage. The numbers on the branches indicate the number of times the sets co-partitioned after 100 iterations.
Figure 6
Linear representations of circular plasmids observed in USA300-HOU-MR (a, b) and USA300-MS (c). Replication and regulatory genes are colored red, recombination/transposition genes are green, antibiotic/heavy metal/bacteriocin resistance genes are blue, other genes are white and hypothetical, conserved hypothetical (CHP) and staphylococcal conserved hypotheticals (SCHP) are colored grey. Horizontal bars indicate the regions of pUSA300-HOU-MR and pUSA300-HOU-MS having homology with other sequenced staphylococcal plasmids.
Figure 7
DNA sequence of the sarU promoters in USA300-HOU-MR and FPR3757 showing -10 and -35 sequences (red) and SarA consensus binding sites (boxed).
Figure 8
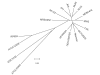
Dendrogram of seventeen fully sequenced genomes generated by Mauve [35] and drawn using MEGA [58]. RP62A and ATCC12228 are S. epidermidis strains, JCSC1435 is an S. haemolyticus strain, and ATCC15305 is an S. saprophyticus strain. All others are S. aureus strains. The bar represents coalescent units.
Similar articles
- Demography and Intercontinental Spread of the USA300 Community-Acquired Methicillin-Resistant Staphylococcus aureus Lineage.
Glaser P, Martins-Simões P, Villain A, Barbier M, Tristan A, Bouchier C, Ma L, Bes M, Laurent F, Guillemot D, Wirth T, Vandenesch F. Glaser P, et al. mBio. 2016 Feb 16;7(1):e02183-15. doi: 10.1128/mBio.02183-15. mBio. 2016. PMID: 26884428 Free PMC article. - Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections.
McCaskill ML, Mason EO Jr, Kaplan SL, Hammerman W, Lamberth LB, Hultén KG. McCaskill ML, et al. Pediatr Infect Dis J. 2007 Dec;26(12):1122-7. doi: 10.1097/INF.0b013e31814536e0. Pediatr Infect Dis J. 2007. PMID: 18043449 - Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus.
Gonzalez BE, Martinez-Aguilar G, Hulten KG, Hammerman WA, Coss-Bu J, Avalos-Mishaan A, Mason EO Jr, Kaplan SL. Gonzalez BE, et al. Pediatrics. 2005 Mar;115(3):642-8. doi: 10.1542/peds.2004-2300. Pediatrics. 2005. PMID: 15741366 - Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus.
Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Diep BA, et al. Lancet. 2006 Mar 4;367(9512):731-9. doi: 10.1016/S0140-6736(06)68231-7. Lancet. 2006. PMID: 16517273 Review. - Treatment of community-acquired methicillin-resistant Staphylococcus aureus in children.
Marcinak JF, Frank AL. Marcinak JF, et al. Curr Opin Infect Dis. 2003 Jun;16(3):265-9. doi: 10.1097/00001432-200306000-00014. Curr Opin Infect Dis. 2003. PMID: 12821819 Review.
Cited by
- A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus.
Kalashnikov M, Lee JC, Campbell J, Sharon A, Sauer-Budge AF. Kalashnikov M, et al. Lab Chip. 2012 Nov 7;12(21):4523-32. doi: 10.1039/c2lc40531h. Lab Chip. 2012. PMID: 22968495 Free PMC article. - Characterizing the transcriptional adaptation of Staphylococcus aureus to stationary phase growth.
Weiss A, Broach WH, Shaw LN. Weiss A, et al. Pathog Dis. 2016 Jul;74(5):ftw046. doi: 10.1093/femspd/ftw046. Epub 2016 May 8. Pathog Dis. 2016. PMID: 27162210 Free PMC article. - rRNA Operon Copy Number Can Explain the Distinct Epidemiology of Hospital-Associated Methicillin-Resistant Staphylococcus aureus.
Fluit AC, Jansen MD, Bosch T, Jansen WT, Schouls L, Jonker MJ, Boel CH. Fluit AC, et al. Antimicrob Agents Chemother. 2016 Nov 21;60(12):7313-7320. doi: 10.1128/AAC.01613-16. Print 2016 Dec. Antimicrob Agents Chemother. 2016. PMID: 27671073 Free PMC article. - Evolutionary paths of streptococcal and staphylococcal superantigens.
Okumura K, Shimomura Y, Murayama SY, Yagi J, Ubukata K, Kirikae T, Miyoshi-Akiyama T. Okumura K, et al. BMC Genomics. 2012 Aug 17;13:404. doi: 10.1186/1471-2164-13-404. BMC Genomics. 2012. PMID: 22900646 Free PMC article. - Molecular Types of Methicillin-Resistant Staphylococcus aureus and Methicillin-Sensitive S. aureus Strains Causing Skin and Soft Tissue Infections and Nasal Colonization, Identified in Community Health Centers in New York City.
Pardos de la Gandara M, Raygoza Garay JA, Mwangi M, Tobin JN, Tsang A, Khalida C, D'Orazio B, Kost RG, Leinberger-Jabari A, Coffran C, Evering TH, Coller BS, Balachandra S, Urban T, Parola C, Salvato S, Jenks N, Wu D, Burgess R, Chung M, de Lencastre H, Tomasz A. Pardos de la Gandara M, et al. J Clin Microbiol. 2015 Aug;53(8):2648-58. doi: 10.1128/JCM.00591-15. Epub 2015 Jun 10. J Clin Microbiol. 2015. PMID: 26063853 Free PMC article.
References
- Coombs GW, Nimmo GR, Bell JM, Huygens F, O'Brien FG, Malkowski MJ, Pearson JC, Stephens AJ, Giffard PM. Genetic diversity amongcommunity methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J Clin Microbiol. 2004;42:4735–4743. doi: 10.1128/JCM.42.10.4735-4743.2004. - DOI - PMC - PubMed
- Chen CJ, Huang YC. Community-acquired methicillin-resistant Staphylococcus aureus in Taiwan. J Microbiol Immunol Infect. 2005;38:376–382. - PubMed
- Hanssen AM, Fossum A, Mikalsen J, Halvorsen DS, Bukholm G, Sollid JU. Dissemination of community-acquired methicillin-resistant Staphylococcus aureus clones in northern Norway: sequence types 8 and 80 predominate. J Clin Microbiol. 2005;43:2118–2124. doi: 10.1128/JCM.43.5.2118-2124.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases