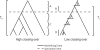Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans - PubMed (original) (raw)
doi: 10.1371/journal.pbio.0050310.
Alisha K Holloway, Kristian Stevens, Ladeana W Hillier, Yu-Ping Poh, Matthew W Hahn, Phillip M Nista, Corbin D Jones, Andrew D Kern, Colin N Dewey, Lior Pachter, Eugene Myers, Charles H Langley
Affiliations
- PMID: 17988176
- PMCID: PMC2062478
- DOI: 10.1371/journal.pbio.0050310
Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans
David J Begun et al. PLoS Biol. 2007.
Abstract
The population genetic perspective is that the processes shaping genomic variation can be revealed only through simultaneous investigation of sequence polymorphism and divergence within and between closely related species. Here we present a population genetic analysis of Drosophila simulans based on whole-genome shotgun sequencing of multiple inbred lines and comparison of the resulting data to genome assemblies of the closely related species, D. melanogaster and D. yakuba. We discovered previously unknown, large-scale fluctuations of polymorphism and divergence along chromosome arms, and significantly less polymorphism and faster divergence on the X chromosome. We generated a comprehensive list of functional elements in the D. simulans genome influenced by adaptive evolution. Finally, we characterized genomic patterns of base composition for coding and noncoding sequence. These results suggest several new hypotheses regarding the genetic and biological mechanisms controlling polymorphism and divergence across the Drosophila genome, and provide a rich resource for the investigation of adaptive evolution and functional variation in D. simulans.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
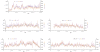
Figure 1. Patterns of Polymorphism and Divergence of Nucleotides along Chromosome Arms
Nucleotide π (blue) and div on the D. simulans lineage (red) in 150-kbp windows are plotted every 10 kbp. χ[–log(p)] (olive) as a measure of deviation (+ or –) in the proportion of polymorphic sites in 30-kbp windows is plotted every 10 kbp (see Materials and Methods). C and T correspond to locations of centromeres and telomeres, respectively. Chromosome arm 3R coordinates correspond to D. simulans locations after accounting for fixed inversion on the D. melanogaster lineage.
Figure 2. Patterns of Polymorphism for Nucleotides, Small Insertions, and Small Deletions along Chromosome Arms
π for nucleotides (blue), π for small (≤ 10 bp) insertions (orange), and π for small (≤ 10 bp) deletions (orchid) among the D. simulans lines in 150-kbp windows are plotted every 10 kbp (see Materials and Methods). C and T correspond to locations of centromeres and telomeres, respectively. Chromosome arm 3R coordinates correspond to D. simulans locations after accounting for fixed inversion on the D. melanogaster lineage.
Figure 3. Rate of Crossing-Over per Base Pair (Green), Nucleotide Polymorphism (Blue) and Nucleotide Divergence (Red) along the X Chromosome
Nucletotide π (blue) and div on the D. simulans lineage (red) in 150-kbp windows are plotted every 10 kbp. Estimated rate of crossing-over (green) is plotted for specific genomic segments (see Materials and Methods).
Figure 4. Hitchhiking Effects Can Induce a Correlation between Polymorphism and Divergence
Hypothetical gene geneoligies in ancestral populations (A or B) and extant populations (C or D) for genomic regions of high crossing-over and low crossing-over (respectively) experiencing different hitchhiking effects. On average, time to the most recent common ancestor in the ancestral population is greater in regions of higher crossing-over (A) and therefore contributes more to the divergence, TH. Regions of lower crossing-over have smaller gene genealogies (D versus C) and less divergence (TL versus TH).
Figure 5. Snapshot of UCSC Browser Tracks in a Genomic Region Showing Significantly Reduced Heterozygosity Relative to Divergence
Nucletotide π (blue, labeled “PI 10K”) and div on the D. simulans lineage (black), labeled “DIV 10K” in 10-kbp windows are plotted every 10 kbp. χ2[-log(p)] (green, labeled “HKA 10K”) as a measure of deviation (+ or −) in the proportion of polymorphic sites in 10-kbp windows is plotted every 10 kbp (see Materials and Methods). The genes scpr-A, scpr-B, and scpr-C exhibit high levels of expression in the testes and are indicated in red.
Similar articles
- Adaptive gene expression divergence inferred from population genomics.
Holloway AK, Lawniczak MK, Mezey JG, Begun DJ, Jones CD. Holloway AK, et al. PLoS Genet. 2007 Oct;3(10):2007-13. doi: 10.1371/journal.pgen.0030187. PLoS Genet. 2007. PMID: 17967066 Free PMC article. - Intron length evolution in Drosophila.
Presgraves DC. Presgraves DC. Mol Biol Evol. 2006 Nov;23(11):2203-13. doi: 10.1093/molbev/msl094. Epub 2006 Aug 21. Mol Biol Evol. 2006. PMID: 16923822 - Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans.
Kern AD, Begun DJ. Kern AD, et al. Genetics. 2008 Jun;179(2):1021-7. doi: 10.1534/genetics.107.078345. Epub 2008 May 27. Genetics. 2008. PMID: 18505885 Free PMC article. - Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.
Kern AD, Begun DJ. Kern AD, et al. Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review. - Historicity and the population genetics of Drosophila melanogaster and D. simulans.
Veuille M, Baudry E, Cobb M, Derome N, Gravot E. Veuille M, et al. Genetica. 2004 Mar;120(1-3):61-70. doi: 10.1023/b:gene.0000017630.69020.32. Genetica. 2004. PMID: 15088647 Review.
Cited by
- Polymorphism-Aware Models in RevBayes: Species Trees, Disentangling Balancing Selection, and GC-Biased Gene Conversion.
Braichenko S, Borges R, Kosiol C. Braichenko S, et al. Mol Biol Evol. 2024 Jul 3;41(7):msae138. doi: 10.1093/molbev/msae138. Mol Biol Evol. 2024. PMID: 38980178 Free PMC article. - Genetic, Environmental, and Stochastic Components of Lifespan Variability: The Drosophila Paradigm.
Bylino OV, Ogienko AA, Batin MA, Georgiev PG, Omelina ES. Bylino OV, et al. Int J Mol Sci. 2024 Apr 19;25(8):4482. doi: 10.3390/ijms25084482. Int J Mol Sci. 2024. PMID: 38674068 Free PMC article. Review. - Shared evolutionary processes shape landscapes of genomic variation in the great apes.
Rodrigues MF, Kern AD, Ralph PL. Rodrigues MF, et al. Genetics. 2024 Apr 3;226(4):iyae006. doi: 10.1093/genetics/iyae006. Genetics. 2024. PMID: 38242701 Free PMC article. - The macronuclear genomic landscape within Tetrahymena thermophila.
Derelle R, Verdonck R, Jacob S, Huet M, Akerman I, Philippe H, Legrand D. Derelle R, et al. Microb Genom. 2024 Jan;10(1):001175. doi: 10.1099/mgen.0.001175. Microb Genom. 2024. PMID: 38206129 Free PMC article. - Whole-genome analysis and evolutionary characterization of cervical and oral human papillomavirus 16.
Minhas S, Kashif M, Nisar H, Idrees M, Ansari F. Minhas S, et al. Exp Biol Med (Maywood). 2023 Dec;248(23):2332-2340. doi: 10.1177/15353702231211861. Epub 2024 Jan 9. Exp Biol Med (Maywood). 2023. PMID: 38196081 Free PMC article.
References
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila . Nature. 1991;351:652–654. - PubMed
- Kimura M. The neutral theory of molecular evolution. Cambridge (UK): Cambridge University Press; 1983.
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG002942/HG/NHGRI NIH HHS/United States
- R01HG2107-3/HG/NHGRI NIH HHS/United States
- R01 GM071926/GM/NIGMS NIH HHS/United States
- R01-HG02362-03/HG/NHGRI NIH HHS/United States
- R01 HG002107/HG/NHGRI NIH HHS/United States
- HG02942-01A1/HG/NHGRI NIH HHS/United States
- R01 HG002362/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases