Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data - PubMed (original) (raw)
Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data
Larry J Wilhelm et al. Biol Direct. 2007.
Abstract
Background: One objective of metagenomics is to reconstruct information about specific uncultured organisms from fragmentary environmental DNA sequences. We used the genome of an isolate of the marine alphaproteobacterium SAR11 ('Candidatus Pelagibacter ubique'; strain HTCC1062), obtained from the cold, productive Oregon coast, as a query sequence to study variation in SAR11 metagenome sequence data from the Sargasso Sea, a warm, oligotrophic ocean gyre.
Results: The average amino acid identity of SAR11 genes encoded by the metagenomic data to the query genome was only 71%, indicating significant evolutionary divergence between the coastal isolates and Sargasso Sea populations. However, an analysis of gene neighbors indicated that SAR11 genes in the Sargasso Sea metagenomic data match the gene order of the HTCC1062 genome in 96% of cases (> 85,000 observations), and that rearrangements are most frequent at predicted operon boundaries. There were no conserved examples of genes with known functions being found in the coastal isolates, but not the Sargasso Sea metagenomic data, or vice versa, suggesting that core regions of these diverse SAR11 genomes are relatively conserved in gene content. However, four hypervariable regions were observed, which may encode properties associated with variation in SAR11 ecotypes. The largest of these, HVR2, is a 48 kb region flanked by the sole 5S and 23S genes in the HTCC1062 genome, and mainly encodes genes that determine cell surface properties. A comparison of two closely related 'Candidatus Pelagibacter' genomes (HTCC1062 and HTCC1002) revealed a number of "gene indels" in core regions. Most of these were found to be polymorphic in the metagenomic data and showed evidence of purifying selection, suggesting that the same "polymorphic gene indels" are maintained in physically isolated SAR11 populations.
Conclusion: These findings suggest that natural selection has conserved many core features of SAR11 genomes across broad oceanic scales, but significant variation was found associated with four hypervariable genome regions. The data also led to the hypothesis that some gene insertions and deletions might be polymorphisms, similar to allelic polymorphisms.
Figures
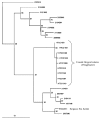
Figure 1
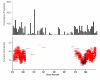
Maximum likelihood tree of recA amino acid sequences. The tree includes Sargasso Sea metagenomic data and predicted proteins from cultured isolates of Pelagibacter. The sequence data was derived from Vergin et al., 2007 [37], with the addition of a sequence from strain HTCC7211, which was recently isolated from the Sargasso Sea. All other HTCC strains are from the coastal Pacific Ocean. The non-HTCC numbers correspond to the fragment identifier in the Sargasso Sea data set. The scale bar indicates substitutions per nucleotide position.
Figure 2
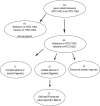
Schematic diagram of procedures used to "bin" the classes of metagenomic data. The query genome, in this case HTCC1062, is represented on the x-axis. A) TBLASTN of protein sequences from the query genome against metagenomic data. "Homologous fragments" were defined as fragments of metagenomic data with expect scores of 1 × 10-10 or better to genes from the query genome. B) "Homlogous fragments with synteny" contain homologs in the same gene order as the query genome, with as many as 5 gene gaps (gene deletions) allowed. C) Best-hit test. Fragments of metagenomic data pass the test if the nucleotide sequence of the fragment gene yields the corresponding query gene as the best hit in a BLASTX search of the NCBI nr database. D) The position of the fragment on the vertical axis corresponds to the average amino acid identity score of all the genes on the fragment.
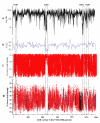
Figure 3
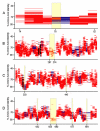
HTCC1062 genome coverage for the different classes of metagenomic data. The data for the figure are described numerically in Table 1. A). The number of homologous fragments (TBLASTX expect scores ≤ 1 × 10-10) for each HTCC1062 gene, plotted by position in the HTCC1062 genome. B) GC content of HTCC1062 genome. C) The distribution of homologous fragments that passed the best-hit test, regardless of synteny. The data in this plot includes fragments that cover one or more genes. The plotted amino acid identities are for the individual genes, not averaged as they are in the syntenic fragment plot below. D) Syntenic fragment coverage of the HTCC1062 genome as a function of gene position and amino acid identity. See Fig. 2 for an explanation of syntenic fragments. Fragments in this category ("bin"), include parts of at least two genes that could be identified by TBLASTX. Regions of blue on the fragments indicate gaps. Syntenic fragments were allowed to be missing as many as five intervening genes (gaps) between the syntenic genes. Genes that encode ribosomal proteins are indicated in black.
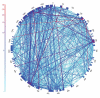
Figure 4
Re-arrangements in the order of SAR11 genes in the Sargasso Sea metagenome, relative to the HTCC1062 genome. The genome of HTCC1062 is represented by the outer circle. Internal lines (chords) indicate SAR11 gene rearrangements found on environmental sequence fragments. The number of occurrences of each gene rearrangement is indicated by the color scale.
Figure 5
Detail of Figs 3C and 3D, in the vicinity of the proteorhodopsin gene. A) homologous fragments that passed the best-hit, and B) syntenic fragments. Regions of blue on the fragments indicate gaps. Only syntenic fragments containing 3 or more genes are shown. (SMRP) small multi-drug resistance protein, (ACAS) acyl-coenzyme A synthetase, (FD) ferrodoxin, (TD) thioredoxin disulfide reductase, (GST) glutathione S-transferase, (DKS) DnaK suppressor protein.
Figure 6
Enlargement of HVR2. HTCC1062 syntenic fragment plot showing detail in the region of HVR2.
Figure 7
Deletion of duplicate genes in the HVR1 region of strains HTCC1002 and HTCC1062. Strain HTCC1002 appears at the top of the display and HTCC1062 at the bottom. One of four homologous Type V autotransporters is deleted in HTCC1062 relative to HTCC1002, and one of two homologous ammonium transporters is deleted in HTCC1002 relative to HTCC1062.
Figure 8
Summary of analysis of HTCC1002/HTCC1062 "gene indels" in core regions. The flow diagram shows how 62 HTCC1002/HTCC1062 gene indels and the data from the syntenic fragment plot were combined to choose 13 genes for the tests of selection data shown in Table 4.
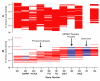
Figure 9
Syntig coverage of HTCC1002/HTCC1062 deletions. Shaded areas highlight the syntig coverage above the genes in HTCC1062 that are deleted in closely related strain HTCC1002. Panels A-G show all 44 genes of this type with HTCC1062 gene numbers on the x-axis.
Similar articles
- A rare SAR11 fosmid clone confirming genetic variability in the 'Candidatus Pelagibacter ubique' genome.
Gilbert JA, Mühling M, Joint I. Gilbert JA, et al. ISME J. 2008 Jul;2(7):790-3. doi: 10.1038/ismej.2008.49. Epub 2008 May 22. ISME J. 2008. PMID: 18496576 - Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade.
Viklund J, Ettema TJ, Andersson SG. Viklund J, et al. Mol Biol Evol. 2012 Feb;29(2):599-615. doi: 10.1093/molbev/msr203. Epub 2011 Sep 7. Mol Biol Evol. 2012. PMID: 21900598 - Proteorhodopsin in the ubiquitous marine bacterium SAR11.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, Rappé MS, Laney S, Wilhelm LJ, Tripp HJ, Mathur EJ, Barofsky DF. Giovannoni SJ, et al. Nature. 2005 Nov 3;438(7064):82-5. doi: 10.1038/nature04032. Nature. 2005. PMID: 16267553 - The unique metabolism of SAR11 aquatic bacteria.
Tripp HJ. Tripp HJ. J Microbiol. 2013 Apr;51(2):147-53. doi: 10.1007/s12275-013-2671-2. Epub 2013 Apr 27. J Microbiol. 2013. PMID: 23625213 Review. - How do divergent ecological strategies emerge among marine bacterioplankton lineages?
Luo H, Moran MA. Luo H, et al. Trends Microbiol. 2015 Sep;23(9):577-84. doi: 10.1016/j.tim.2015.05.004. Epub 2015 Jun 4. Trends Microbiol. 2015. PMID: 26051014 Review.
Cited by
- Global biogeography of SAR11 marine bacteria.
Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, Riddle MJ, Fuhrman JA, Andrews-Pfannkoch C, Hoffman JM, McQuaid JB, Allen A, Rintoul SR, Cavicchioli R. Brown MV, et al. Mol Syst Biol. 2012 Jul 17;8:595. doi: 10.1038/msb.2012.28. Mol Syst Biol. 2012. PMID: 22806143 Free PMC article. - Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds.
Oh D, Porter K, Russ B, Burns D, Dyall-Smith M. Oh D, et al. Extremophiles. 2010 Mar;14(2):161-9. doi: 10.1007/s00792-009-0295-6. Epub 2009 Dec 20. Extremophiles. 2010. PMID: 20091074 Free PMC article. - Reanalyze unassigned reads in Sanger based metagenomic data using conserved gene adjacency.
Weng FC, Su CH, Hsu MT, Wang TY, Tsai HK, Wang D. Weng FC, et al. BMC Bioinformatics. 2010 Nov 18;11:565. doi: 10.1186/1471-2105-11-565. BMC Bioinformatics. 2010. PMID: 21083935 Free PMC article. - The analysis of groEL gene in Salmonella enterica subspecies enterica serovar Typhimurium isolated from avians by PCR-Restriction Fragment Length Polymorphism method.
Dilmaghani M, Ahmadi M, Zahraei Salehi T, Talebi A. Dilmaghani M, et al. Vet Res Commun. 2011 Mar;35(3):133-43. doi: 10.1007/s11259-011-9460-3. Epub 2011 Feb 11. Vet Res Commun. 2011. PMID: 21312060 - Tuning fresh: radiation through rewiring of central metabolism in streamlined bacteria.
Eiler A, Mondav R, Sinclair L, Fernandez-Vidal L, Scofield DG, Schwientek P, Martinez-Garcia M, Torrents D, McMahon KD, Andersson SG, Stepanauskas R, Woyke T, Bertilsson S. Eiler A, et al. ISME J. 2016 Aug;10(8):1902-14. doi: 10.1038/ismej.2015.260. Epub 2016 Jan 19. ISME J. 2016. PMID: 26784354 Free PMC article.
References
- Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. - DOI - PMC - PubMed
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources