Cell division modulates prion accumulation in cultured cells - PubMed (original) (raw)
Cell division modulates prion accumulation in cultured cells
Sina Ghaemmaghami et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The phenotypic effect of prions on host cells is influenced by the physical properties of the prion strain and its level of accumulation. In mammalian cell cultures, prion accumulation is determined by the interplay between de novo prion formation, catabolism, cell division, and horizontal cell-to-cell transmission. Understanding this dynamic enables the analytical modeling of protein-based heritability and infectivity. Here, we quantitatively measured these competing effects in a subline of neuroblastoma (N2a) cells and propose a concordant reaction mechanism to explain the kinetics of prion propagation. Our results show that cell division leads to a predictable reduction in steady-state prion levels but not to complete clearance. Scrapie-infected N2a cells were capable of accumulating different steady-state levels of prions, dictated partly by the rate of cell division. We also show that prions in this subline of N2a cells are transmitted primarily from mother to daughter cells, rather than horizontal cell-to-cell transmission. We quantitatively modeled our kinetic results based on a mechanism that assumes a subpopulation of prions is capable of self-catalysis, and the levels of this subpopulation reach saturation in fully infected cells. Our results suggest that the apparent effectiveness of antiprion compounds in culture may be strongly influenced by the growth phase of the target cells.
Conflict of interest statement
Conflict of interest statement: B.C.H.M., F.E.C., and S.B.P. have a financial interest in InPro Biotechnology, Inc.
Figures
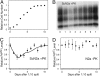
Fig. 1.
Cell division decreases the intracellular accumulation of PrPSc but not PrPC. (A) Growth curve for ScN2a cells after a 1:10 split in 100-mm plates. (B–D) The change in the accumulation of PrPSc in ScN2a (B and C) and PrPC in N2a (D) as cells progress from a logarithmic growth phase to stationary phase, as detected by Western blot (B) and ELISA (C and D). After lysis, all samples were normalized to 1 mg/ml total protein concentration as determined by the BCA assay (Pierce) before further processing. The plotted ELISA measurements were normalized to measurements at time 0. The dashed lines indicate the background signal level as determined by plating a mock PBS + 1% BSA sample. The error bars represent standard deviations of three replicate measurements. The solid line in C indicates the predicted PrPSc fluctuation based on the limited conversion model and Eq. 8 (
SI Text
).
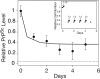
Fig. 2.
Dividing cells are able to maintain a reduced steady-state level of PrPSc. ELISA measurements of PrPSc levels were normalized to measurements at time 0. The dashed line indicates a background signal from mock PBS + 1% BSA. The solid line represents predicted PrPSc levels according to the limited conversion model and Eq. 8 (
SI Text
). Error bars indicate standard deviations of three replicate measurements. Inset shows the experimental design. ScN2a cells were maintained in a perpetual state of cell division for 5 days by repetitive cycles of subconfluent dissociation and replating (see Materials and Methods). The arrows represent the times of dissociation and replating at the indicated ratios. The predicted cell numbers (relative to a confluent culture) during the course of the experiment are also shown.
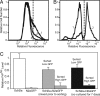
Fig. 3.
In ScN2a cultures, the rate of horizontal cell-to-cell infection is slow relative to the rate of cell division. (A) Fluorescence distribution in the following cell lines, as measured by FACS: (1) N2a cells stably transfected with GFP, (2) a 7-day coculture consisting of a 1:1 ratio of ScN2a-N2aGFP cells, and (3) nontransfected ScN2a cells. The dashed line indicates the gate used to separate “high GFP” cells from “low GFP” cells. (B) After FACS, the fluorescence distribution of the sorted coculture shows that “high GFP” subpopulation (2) was successfully separated from “low GFP” subpopulation (1). (C) PrPSc levels of the sorted subpopulations from 1:1 Sc2Na−N2aGFP mixtures, as determined by ELISA. PrPSc levels from sorted subpopulations are similar, whether the ScN2a + N2aGFP mixtures were cultured independently then mixed before FACS analysis (gray bars) or cocultured for 7 days (black bars). These data indicate that cells descending from N2aGFP do not contain levels of PrPSc above the experimental error of the experiment. As controls, PrPSc levels in 100% ScN2a and N2aGFP cultures (white bars) are plotted. The PrPSc signal in the sorted low GFP population is reduced because of the presence of transfected N2a cells with low GFP fluorescence levels (see profile 1 in A). The PrPSc signal seen in the sorted high GFP population is due to cross-contamination and represents the experimental background because of sorting artifacts.
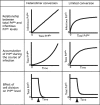
Fig. 4.
Features and predictions of the limited conversion model compared with the simplified heterodimer conversion model. The limited conversion model posits that in fully infected cultures, when the cell accumulates high concentrations of PrPSc, the subpopulation of the PrPSc that is infectious (capable of self-catalysis) reaches saturation levels (Top). This assumption alleviates some of the experimental inconsistencies associated with treating the processes of PrPSc conversion, PrPSc degradation, and cell division as first-order reactions. The model explains the ability of cells in culture to attain stable levels of PrPSc when fully infected (Middle). It also provides an explanation for the ability of cells to achieve different steady-state levels of PrPSc when dividing at different rates (Bottom). In the simulation shown, stationary cells that have achieved a steady-state level of PrPSc are induced to divide at a time indicated by the arrow. In such a scenario, the simplified heterodimer model predicts a complete clearance of PrPSc over time, whereas the limited conversion model predicts a new steady-state level dictated by Eq. 6. The heterodimer model predicts an unstable state when [PrPC] is assumed to remain constant (as has been experimentally shown here). When this constraint is removed, the model is able to predict a stable state.
Similar articles
- Prions amplify through degradation of the VPS10P sorting receptor sortilin.
Uchiyama K, Tomita M, Yano M, Chida J, Hara H, Das NR, Nykjaer A, Sakaguchi S. Uchiyama K, et al. PLoS Pathog. 2017 Jun 30;13(6):e1006470. doi: 10.1371/journal.ppat.1006470. eCollection 2017 Jun. PLoS Pathog. 2017. PMID: 28665987 Free PMC article. - Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles.
Alais S, Simoes S, Baas D, Lehmann S, Raposo G, Darlix JL, Leblanc P. Alais S, et al. Biol Cell. 2008 Oct;100(10):603-15. doi: 10.1042/BC20080025. Biol Cell. 2008. PMID: 18422484 - Cholesterol transporter ATP-binding cassette A1 (ABCA1) is elevated in prion disease and affects PrPC and PrPSc concentrations in cultured cells.
Kumar R, McClain D, Young R, Carlson GA. Kumar R, et al. J Gen Virol. 2008 Jun;89(Pt 6):1525-1532. doi: 10.1099/vir.0.83358-0. J Gen Virol. 2008. PMID: 18474570 Free PMC article. - Cell culture models of transmissible spongiform encephalopathies.
Béranger F, Mangé A, Solassol J, Lehmann S. Béranger F, et al. Biochem Biophys Res Commun. 2001 Nov 30;289(2):311-6. doi: 10.1006/bbrc.2001.5941. Biochem Biophys Res Commun. 2001. PMID: 11716473 Review. - Prion protein conversion in vitro.
Supattapone S. Supattapone S. J Mol Med (Berl). 2004 Jun;82(6):348-56. doi: 10.1007/s00109-004-0534-3. Epub 2004 Mar 10. J Mol Med (Berl). 2004. PMID: 15014886 Review.
Cited by
- Cell-to-cell propagation of infectious cytosolic protein aggregates.
Hofmann JP, Denner P, Nussbaum-Krammer C, Kuhn PH, Suhre MH, Scheibel T, Lichtenthaler SF, Schätzl HM, Bano D, Vorberg IM. Hofmann JP, et al. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5951-6. doi: 10.1073/pnas.1217321110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509289 Free PMC article. - Effect of Scrapie Prion Infection in Ovine Bone Marrow-Derived Mesenchymal Stem Cells and Ovine Mesenchymal Stem Cell-Derived Neurons.
García-Mendívil L, Mediano DR, Hernaiz A, Sanz-Rubio D, Vázquez FJ, Marín B, López-Pérez Ó, Otero A, Badiola JJ, Zaragoza P, Ordovás L, Bolea R, Martín-Burriel I. García-Mendívil L, et al. Animals (Basel). 2021 Apr 15;11(4):1137. doi: 10.3390/ani11041137. Animals (Basel). 2021. PMID: 33921147 Free PMC article. - Proliferative arrest of neural cells induces prion protein synthesis, nanotube formation, and cell-to-cell contacts.
Miyazawa K, Emmerling K, Manuelidis L. Miyazawa K, et al. J Cell Biochem. 2010 Sep 1;111(1):239-47. doi: 10.1002/jcb.22723. J Cell Biochem. 2010. PMID: 20518071 Free PMC article. - Discovery of 2-aminothiazoles as potent antiprion compounds.
Ghaemmaghami S, May BC, Renslo AR, Prusiner SB. Ghaemmaghami S, et al. J Virol. 2010 Apr;84(7):3408-12. doi: 10.1128/JVI.02145-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032192 Free PMC article. - Propagation of prions causing synucleinopathies in cultured cells.
Woerman AL, Stöhr J, Aoyagi A, Rampersaud R, Krejciova Z, Watts JC, Ohyama T, Patel S, Widjaja K, Oehler A, Sanders DW, Diamond MI, Seeley WW, Middleton LT, Gentleman SM, Mordes DA, Südhof TC, Giles K, Prusiner SB. Woerman AL, et al. Proc Natl Acad Sci U S A. 2015 Sep 1;112(35):E4949-58. doi: 10.1073/pnas.1513426112. Epub 2015 Aug 18. Proc Natl Acad Sci U S A. 2015. PMID: 26286986 Free PMC article.
References
- Prusiner SB. N Engl J Med. 2001;344:1516–1526. - PubMed
- Basler K, Oesch B, Scott M, Westaway D, Wälchli M, Groth DF, McKinley MP, Prusiner SB, Weissmann C. Cell. 1986;46:417–428. - PubMed
- Pan YT, Hori H, Saul R, Sanford BA, Molyneux RJ, Elbein AD. Biochemistry. 1983;22:3975–3984. - PubMed
- Prusiner SB. Annu Rev Med. 1987;38:381–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG002132/AG/NIA NIH HHS/United States
- P01 AG010770/AG/NIA NIH HHS/United States
- P01 AG021601/AG/NIA NIH HHS/United States
- AG021601/AG/NIA NIH HHS/United States
- AG02132/AG/NIA NIH HHS/United States
- AG10770/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources