Examining protein protein interactions using endogenously tagged yeast arrays: the cross-and-capture system - PubMed (original) (raw)
. 2007 Dec;17(12):1774-82.
doi: 10.1101/gr.6667007. Epub 2007 Nov 7.
Michael J Fetchko, Ralph Imhof, Christopher I Graham, Ingrid Stoffel-Studer, Caroline Zbinden, Maanasa Raghavan, Lianet Lopez, Lucija Beneti, Jacqueline Hort, Jeffrey Fillingham, Jack F Greenblatt, Guri Giaever, Corey Nislow, Igor Stagljar
Affiliations
- PMID: 17989249
- PMCID: PMC2099586
- DOI: 10.1101/gr.6667007
Examining protein protein interactions using endogenously tagged yeast arrays: the cross-and-capture system
Bernhard Suter et al. Genome Res. 2007 Dec.
Abstract
Comprehensive approaches to detect protein-protein interactions (PPIs) have been most successful in the yeast model system. Here we present "Cross-and-Capture," a novel assay for rapid, sensitive assessment of PPIs via pulldown of differently tagged yeast strain arrays. About 500 yeast genes that function in DNA replication, repair, and recombination and nuclear proteins of unknown function were chromosomally tagged with six histidine residues or triple VSV epitopes. We demonstrate that the assay can interrogate a wide range of previously known protein complexes with increased resolution and sensitivity. Furthermore, we use "Cross-and-Capture" to identify two novel protein complexes: Rtt101p-Mms1p and Sae2p-Mre11p. The Rtt101p-Mms1p interaction was subsequently characterized by genetic and functional analyses. Our studies establish the "Cross-and-Capture" assay as a novel, versatile tool that provides a valuable complement for the next generation of yeast proteomic studies.
Figures
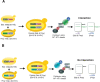
Figure 1.
Cross-and-Capture Assay. (A,B) A strain containing the bait ORFX tagged with a V5 epitope and six histidines (6×HIS) is crossed with strains that contain prey ORFY or ORFZ tagged with a V5 epitope followed by a triple VSV tag (3×VSV). Diploids, which express both tagged bait and prey, are grown on selective medium. Protein extracts from the diploids are then incubated with nickel beads (Ni2+-NTA), allowing isolation of bait (Protein X–6×HIS) and bait-associated prey protein (Protein Y–3×VSV) (A), whereas a non-interacting protein (Protein Z–3×VSV) will not bind (B). Proteins are then separated by SDS-PAGE, and blots are probed for bait and prey (anti-V5 antibody) and specifically for the prey (anti-VSV antibody) by immunoblot.
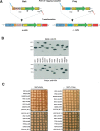
Figure 2.
Generation and verification of tagged protein arrays. (A) To tag ORFX as bait (V5–6×HIS) and prey (V5–3×VSV), a set of primers is used that anneal to identical binding sites within the template plasmids and have flanking sequence homologous to ORFX. PCR products generated from the bait and prey templates are transformed into a- and α-cells, respectively. Homologous recombination occurs between the variable portion of the 5′ primer (light blue) and the 3′ terminus of the ORF, and between the variable portion of the 3′ primer (red) and the 3′ UTR) of ORFX. Transformants are selected on G418 plates, and colony PCR is performed to verify integration of the _Kan_r downstream of the desired ORF. Abbreviations: TEF, translational elongation factor; TEFp, TEF promoter; TEFt, TEF terminator: _Kan_r, kanamycin resistance; loxp, site for CRE specific homologous recombination. (B) Monitoring the quality of the arrays by Western analysis of 14 ORFs as baits and preys. We chose proteins with a high level of expression (>2000 molecules per cell) as judged by Ghaemmaghami et al. (2003). The asterisk (*) denotes possible misloading or protein degradation. Note in the RAD51 lane the multiple protein products. Expected protein sizes are listed in Supplemental Table 1. (C) Analysis of effects on cell growth by tagging essential genes. A total of 24 strains with essential genes tagged as baits (6×HIS) and preys (3×VSV) were grown to saturation and spotted in 10-fold dilutions on YPD. Pictures were taken after 2 d at 30°C.
Figure 3.
Proof-of-principle of Cross-and-Capture. (A) Detection of known protein complexes with Cross-and-Capture. Two diploids were examined, one expressing the bait and prey and the other expressing only the prey. Whole-cell extracts (WCEs) and pulldowns are probed with anti-V5. Pulldowns are additionally probed with anti-VSV. The origin-recognition complex (Orc1p–Orc5p), the chromatin assembly complex (Cac2p–Rlf2p), the casein kinase complex (Cka2p–Cka1p), DNA primase (Pri1p–Pri2p), the cyclin/TFIIH complex (Ccl1p–Tfb1p), and the Rad53p–Asf1p interaction are shown. For Rad53p–Asf1p interaction, growth for 1 h in 3 μg/mL 4NQO, is indicated by “+”. Note the band shift by phosphorylation (Rad53P–3×VSV) in presence of 4NQO. (B) Identification of the Shu1p–Psy3p complex. Pulldowns are done from diploids expressing Psy3 and Shu1 as both baits and preys. WCEs and pulldown are probed with anti-V5 and anti-VSV antibodies. Nonspecific background signals in the Western blots are indicated (*).
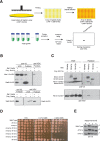
Figure 4.
Using Cross-and-Capture to screen for novel PPIs. (A) Screening scheme. A strain containing a prey (_ORFY_–3×VSV) is combined with a set of different bait strains (_ORF_1-506–6×HIS) and diploids are selected. Extracts and Western blots are processed in parallel, and pulldowns of the prey protein are assessed. (B) Sae2p interacts with Mre11p and Sae2p. Pulldowns are from diploids expressing Mre11–3×VSV or Sae2–3×VSV as prey and Sae2–6×HIS as bait or no bait control (−). (C) Identification of the Rtt101p–Mms1p complex. Whole cell extracts (WCEs) and pulldowns probed with anti-V5 and anti-VSV antibodies are shown. Cells are treated with 0.02% MMS for 2 h before harvesting. Pulldowns are from diploids expressing Rtt101–3×VSV as prey and Mms1p, Mms4p, and Rtt107p as bait proteins (6×HIS) or no bait control (−). Nonspecific background signals in the Western blots are indicated (*). (D) Epistasis analysis for _rtt101_Δ and _mms1_Δ compared to _rtt107_Δ combined with _mms1_Δ. Cells are grown to mid-log phase and analyzed by spot assay in fivefold serial dilutions on YPD, YPD + 0.01% MMS, and YPD + 0.02% MMS. Pictures were taken after 2–3 d at 30°C. (E) Phosphorylation status of Rad53p (P) was determined by blotting Rad53–6×HIS from wild type, _rtt101_Δ, and _mms1_Δ strains with anti-V5 at indicated time points. Cells are arrested in G1 for 2 h with α-factor (α) and released into YPD containing 0.033% MMS for 2 h (+). Cells are then incubated with fresh YPD, and samples are collected after 2 and 4 h.
Similar articles
- The Cross-and-Capture system: a versatile tool in yeast proteomics.
Suter B. Suter B. Methods. 2012 Dec;58(4):360-6. doi: 10.1016/j.ymeth.2012.07.017. Epub 2012 Jul 24. Methods. 2012. PMID: 22836129 - Exploring protein phosphorylation in response to DNA damage using differentially tagged yeast arrays.
Suter B, Graham C, Stagljar I. Suter B, et al. Biotechniques. 2008 Nov;45(5):581-4. doi: 10.2144/000112949. Biotechniques. 2008. PMID: 19007343 - PIPE: a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs.
Pitre S, Dehne F, Chan A, Cheetham J, Duong A, Emili A, Gebbia M, Greenblatt J, Jessulat M, Krogan N, Luo X, Golshani A. Pitre S, et al. BMC Bioinformatics. 2006 Jul 27;7:365. doi: 10.1186/1471-2105-7-365. BMC Bioinformatics. 2006. PMID: 16872538 Free PMC article. - [Decade of genomics--methods for genome investigation in yeast Saccharomyces cerevisiae].
Skoneczna A. Skoneczna A. Postepy Biochem. 2006;52(4):435-47. Postepy Biochem. 2006. PMID: 17536513 Review. Polish. - Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond.
Suter B, Auerbach D, Stagljar I. Suter B, et al. Biotechniques. 2006 May;40(5):625-44. doi: 10.2144/000112151. Biotechniques. 2006. PMID: 16708762 Review.
Cited by
- Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA.
Zaidi IW, Rabut G, Poveda A, Scheel H, Malmström J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Zaidi IW, et al. EMBO Rep. 2008 Oct;9(10):1034-40. doi: 10.1038/embor.2008.155. Epub 2008 Aug 15. EMBO Rep. 2008. PMID: 18704118 Free PMC article. - A role for ubiquitin in the clearance of nonfunctional rRNAs.
Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. Fujii K, et al. Genes Dev. 2009 Apr 15;23(8):963-74. doi: 10.1101/gad.1775609. Genes Dev. 2009. PMID: 19390089 Free PMC article. - Identifying novel protein complexes in cancer cells using epitope-tagging of endogenous human genes and affinity-purification mass spectrometry.
Song J, Hao Y, Du Z, Wang Z, Ewing RM. Song J, et al. J Proteome Res. 2012 Dec 7;11(12):5630-41. doi: 10.1021/pr300598t. Epub 2012 Nov 7. J Proteome Res. 2012. PMID: 23106643 Free PMC article. - Active destruction of defective ribosomes by a ubiquitin ligase involved in DNA repair.
Hinnebusch AG. Hinnebusch AG. Genes Dev. 2009 Apr 15;23(8):891-5. doi: 10.1101/gad.1800509. Genes Dev. 2009. PMID: 19390082 Free PMC article. - The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes.
Vincent NG, Charette JM, Baserga SJ. Vincent NG, et al. RNA. 2018 Jan;24(1):77-89. doi: 10.1261/rna.062927.117. Epub 2017 Oct 20. RNA. 2018. PMID: 29054886 Free PMC article.
References
- Bell S.P., Dutta A., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. - PubMed
- Clerici M., Mantiero D., Lucchini G., Longhese M.P., Mantiero D., Lucchini G., Longhese M.P., Lucchini G., Longhese M.P., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. - PubMed
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Schuldiner M., Gebbia M., Recht J., Shales M., Gebbia M., Recht J., Shales M., Recht J., Shales M., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases