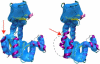Fitting low-resolution cryo-EM maps of proteins using constrained geometric simulations - PubMed (original) (raw)
Fitting low-resolution cryo-EM maps of proteins using constrained geometric simulations
Craig C Jolley et al. Biophys J. 2008.
Erratum in
- Biophys J. 2008 Apr 15;94(8):3361
Abstract
Recent experimental advances in producing density maps from cryo-electron microscopy (cryo-EM) have challenged theorists to develop improved techniques to provide structural models that are consistent with the data and that preserve all the local stereochemistry associated with the biomolecule. We develop a new technique that maintains the local geometry and chemistry at each stage of the fitting procedure. A geometric simulation is used to drive the structure from some appropriate starting point (a nearby experimental structure or a modeled structure) toward the experimental density, via a set of small incremental motions. Structural motifs such as alpha-helices can be held rigid during the fitting procedure as the starting structure is brought into alignment with the experimental density. After validating this procedure on simulated data for adenylate kinase and lactoferrin, we show how cryo-EM data for two different GroEL structures can be fit using a starting x-ray crystal structure. We show that by incorporating the correct local stereochemistry in the modeling, structures can be obtained with effective resolution that is significantly higher than might be expected from the nominal cryo-EM resolution.
Figures
Figure 1
Atomic electron densities used to generate simulated cryo-EM data. The calculated electron densities for carbon, nitrogen, oxygen, and sulfur are shown. These are the “zero-resolution” densities; simulated finite-resolution data is obtained by convolving each of these with a Gaussian function.
Figure 2
(A) Fitting of ADK into a theoretical density map with 4 Å resolution. The map was generated using the open form of ADK (PDB ID 4AKE), and the image on the left shows the best rigid fit (using FRODA) between this map and the closed form of ADK (PDB ID 1AKE). The image on the right shows the result of flexible fitting. (B) Fitting of lactoferrin into a theoretic map with 5 Å resolution. The map was generated using the open form of lactoferrin (PDB ID 1LFH), and the simulation was started from the closed form (PDB ID 1LFG). (C) Fitting of GroEL into an experimental map at 14.9 Å resolution. The images shown are from the intact domain fitting (see text for details). The Supplementary Material accompanying this article contains animated versions of these trajectories.
Figure 3
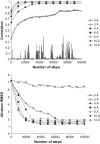
Plots of the real-space correlation (top) and all-atom RMSD (bottom) for results of ADK fitting with resolutions from 0–13.3 Å using a 1 Å grid. Some curves have been omitted for visual clarity. Note that the curves for both the correlation and the RMSD reach a steady asymptotic value at the same point, around conformer 30,000. The RMSD plot shows that the fitting for resolutions >2.0 Å was successful, whereas the higher-resolution simulations generally failed to converge. The 0 Å simulation shows a lack of any clear convergence; the spiky appearance of the 0 Å correlation plot results from high noise sensitivity at small values of the correlation. Complete plots for 1–4 Å grids are contained in the Supplementary Material accompanying this article.
Figure 4
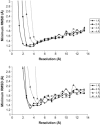
The relationship between resolution and fit quality (measured by the minimum RMSD between the simulated and target structures) for ADK and lactoferrin. Notice that the fit quality deteriorates rapidly as the grid spacing approaches the resolution length scale, and that the choice of grid spacing seems to be unimportant as long as it is smaller than the length scale imposed by resolution. For grid spacings that gave an acceptable fit, there is a roughly linear relationship in which fit quality improves with resolution.
Figure 5
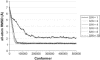
The effects of noise on ADK fitting at 3.3 Å resolution. The simulations converged for all levels of noise considered, although the quality of the fit was diminished for extremely high levels of signal/noise (S/N = 1). The results seem to be nearly identical for signal/noise ratios >1, indicating that this flexible fitting method is robust in the presence of relatively high levels of noise.
Figure 6
Effects of missing density on ADK fitting at 8.0 Å resolution. (Left) Results of fitting the closed structure of ADK (Fig. 1) to a map generated using the complete open structure of ADK. (Right) Before the map was generated, the C-terminal helix (indicated with a red arrow) was deleted, resulting in missing density, marked with a dotted line in the image on the right. When the complete closed form of ADK was fit into this incomplete map, the “extra” C-terminal helix was pushed upward into the density, resulting in some distortion of the surrounding region. Regions of the structure relatively distant from the modification are largely unchanged.
Figure 7
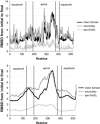
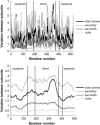
RMSD between initial and final structures for each GroEL fitting trajectory, averaged over the 14 subunits in the complex. The lower plot shows the same data as the upper plot, except that the curve was smoothed by averaging the RMSD values for 50 adjacent residues at each point. For each simulation, the region of largest mobility is the apical domain, with smaller motions in the intermediate domain. Divisions between the domains are denoted with vertical lines.
Figure 8
Average deviation between the different subunits within a given fitted structure. The data in the lower plot is smoothed as in Fig. 7. The largest variance is in the apical and intermediate domains (Fig. 7), consistent with the observation that the regions that experience the largest motion also show the largest variance in their final positions. Vertical lines show the division between the equatorial, intermediate, and apical domains.
Similar articles
- Docking structures of domains into maps from cryo-electron microscopy using local correlation.
Roseman AM. Roseman AM. Acta Crystallogr D Biol Crystallogr. 2000 Oct;56(Pt 10):1332-40. doi: 10.1107/s0907444900010908. Acta Crystallogr D Biol Crystallogr. 2000. PMID: 10998630 - Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy.
Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Ludtke SJ, et al. Structure. 2004 Jul;12(7):1129-36. doi: 10.1016/j.str.2004.05.006. Structure. 2004. PMID: 15242589 - Refinement of Atomic Structures Against cryo-EM Maps.
Murshudov GN. Murshudov GN. Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review. - High-resolution modeling of protein structures based on flexible fitting of low-resolution structural data.
Zheng W, Tekpinar M. Zheng W, et al. Adv Protein Chem Struct Biol. 2014;96:267-84. doi: 10.1016/bs.apcsb.2014.06.004. Epub 2014 Aug 24. Adv Protein Chem Struct Biol. 2014. PMID: 25443961 Review.
Cited by
- ATTRACT-EM: a new method for the computational assembly of large molecular machines using cryo-EM maps.
de Vries SJ, Zacharias M. de Vries SJ, et al. PLoS One. 2012;7(12):e49733. doi: 10.1371/journal.pone.0049733. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23251350 Free PMC article. - Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations.
Orzechowski M, Tama F. Orzechowski M, et al. Biophys J. 2008 Dec 15;95(12):5692-705. doi: 10.1529/biophysj.108.139451. Epub 2008 Oct 10. Biophys J. 2008. PMID: 18849406 Free PMC article. - Computational Methodologies for Real-Space Structural Refinement of Large Macromolecular Complexes.
Goh BC, Hadden JA, Bernardi RC, Singharoy A, McGreevy R, Rudack T, Cassidy CK, Schulten K. Goh BC, et al. Annu Rev Biophys. 2016 Jul 5;45:253-78. doi: 10.1146/annurev-biophys-062215-011113. Epub 2016 May 2. Annu Rev Biophys. 2016. PMID: 27145875 Free PMC article. Review. - Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics.
Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Trabuco LG, et al. Structure. 2008 May;16(5):673-83. doi: 10.1016/j.str.2008.03.005. Structure. 2008. PMID: 18462672 Free PMC article. - Variability of Protein Structure Models from Electron Microscopy.
Monroe L, Terashi G, Kihara D. Monroe L, et al. Structure. 2017 Apr 4;25(4):592-602.e2. doi: 10.1016/j.str.2017.02.004. Epub 2017 Mar 2. Structure. 2017. PMID: 28262392 Free PMC article.
References
- Baker T.S., Johnson J.E. Low resolution meets high: towards a resolution continuum from cells to atoms. Curr. Opin. Struct. Biol. 1996;6:585–594. - PubMed
- Rossmann M.G. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1341–1349. - PubMed
- Fabiola F., Chapman M.S. Fitting of high-resolution structures into electron microscopy reconstruction images. Structure. 2005;13:389–400. - PubMed
- Wriggers W., Birmanns S. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 2001;133:193–202. - PubMed
- Chen J.Z., Furst J., Chapman M.S., Grigorieff N. Low-resolution structure refinement in electron microscopy. J. Struct. Biol. 2003;144:144–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials