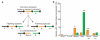Demasculinization of X chromosomes in the Drosophila genus - PubMed (original) (raw)
Demasculinization of X chromosomes in the Drosophila genus
David Sturgill et al. Nature. 2007.
Abstract
X chromosomes evolve differently from autosomes, but general governing principles have not emerged. For example, genes with male-biased expression are under-represented on the X chromosome of D. melanogaster, but are randomly distributed in the genome of Anopheles gambiae. In direct global profiling experiments using species-specific microarrays, we find a nearly identical paucity of genes with male-biased expression on D. melanogaster, D. simulans, D. yakuba, D. ananassae, D. virilis and D. mojavensis X chromosomes. We observe the same under-representation on the neo-X of D. pseudoobscura. It has been suggested that precocious meiotic silencing of the X chromosome accounts for reduced X chromosome male-biased expression in nematodes, mammals and Drosophila. We show that X chromosome genes with male-biased expression are under-represented in somatic cells and in mitotic male germ cells. These data are incompatible with simple X chromosome inactivation models. Using expression profiling and comparative sequence analysis, we show that selective gene extinction on the X chromosome, creation of new genes on autosomes and changed genomic location of existing genes contribute to the unusual X chromosome gene content.
Figures
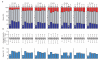
Figure 1. Genes that show sex-biased expression, on chromosome arms
a, Percentage of genes with female-biased (red), non-biased (grey) and male-biased (blue) expression on chromosome arms. Muller's elements and arms are indicated (X chromosomes in bold). Significant under-representation from a random distribution of genes in an expression class (chi-squared test) is noted (*P<10−4). The top 20% of differentially expressed genes (by ranked P values) are assigned a sex-biased expression class. b, Box plots of average hybridization intensities for males by chromosome arm. Twenty-fifth to seventy fifth percentiles (boxes), medians (lines in boxes) and ranges (whiskers) are indicated. c, As in a, but with genes with predicted testis-biased expression removed. D. melanogaster, D. mel; D. simulans, D. sim, D. yakuba, D. yak; D. ananassae, D. ana; D. pseudoobscura, D. pse; D. virilis, D. vir; D. mojavensis, D. moj.
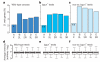
Figure 2. Male-biased expression not subject to X chromosome inactivation
a–c, Percentage of D. melanogaster genes with male-biased expression. Significant under-representation as in Fig. 1 is noted (*P<10−2; **P<10−4). a, Non-gonadal soma (gonadectomized carcasses). b, c, Testis from mutants with mitotically active germline tumours. Male-biased expression was determined by present calls in wild-type and mutant testis and absent calls in wild-type ovary expression profiles from FlyMine. Testis from _bgcn_− males (primary spermatocyte-biased; b) accumulates mitotically active cysts of interconnected germ cells, whereas testis from males overexpressing os (nos-os) in a _bgcn_− background (male germline stem-cell-biased; c) accumulates mitotically active germ cells that undergo complete cytokinesis. d–f, Box plots of average intensities (arbitrary scale) across hybridizations for wild-type (d), _bgcn_− (e), or _nos-os; bgcn_− (f) testis separated by chromosome arm. Twenty-fifth to seventy-fifth percentiles (boxes), medians (lines in boxes) and ranges (whiskers) are indicated.
Figure 3. The neo-X chromosome
a, D. pseudoobscura neo-X chromosome formation cartoon. Muller A, D and E are attached to different centromeres (filled circles) in D. pseudoobscura relative to flanking species. b, Bar plot showing changes to D. pseudoobscura Muller D neo-X chromosome (orange) and Muller E autosomal arm (green), based on the predicted expression pattern of the ancestral autosomes. Categories are: switches in sex-biased expression (F, female-biased; M, male-biased), not found (absent), only found in D. pseudoobscura (new), and genes moving from (move off) and to (move on) the indicated arm. Significant differences (chi-squared test) between Muller D and Muller E are noted (**P<10−4; *P<0.05). The ancestral condition of the neo-X chromosome was inferred by examining chromosome linkage and expression on homologous arms in species that diverged from the Drosophila lineage before and after D. pseudoobscura.
Similar articles
- Demasculinization of the Anopheles gambiae X chromosome.
Magnusson K, Lycett GJ, Mendes AM, Lynd A, Papathanos PA, Crisanti A, Windbichler N. Magnusson K, et al. BMC Evol Biol. 2012 May 18;12:69. doi: 10.1186/1471-2148-12-69. BMC Evol Biol. 2012. PMID: 22607633 Free PMC article. - Constraint and turnover in sex-biased gene expression in the genus Drosophila.
Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Zhang Y, et al. Nature. 2007 Nov 8;450(7167):233-7. doi: 10.1038/nature06323. Nature. 2007. PMID: 17994089 Free PMC article. - Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome.
Vibranovski MD, Zhang YE, Kemkemer C, Lopes HF, Karr TL, Long M. Vibranovski MD, et al. BMC Biol. 2012 Jun 12;10:49; author reply 50. doi: 10.1186/1741-7007-10-49. BMC Biol. 2012. PMID: 22691264 Free PMC article. - Gene content evolution on the X chromosome.
Gurbich TA, Bachtrog D. Gurbich TA, et al. Curr Opin Genet Dev. 2008 Dec;18(6):493-8. doi: 10.1016/j.gde.2008.09.006. Epub 2008 Oct 16. Curr Opin Genet Dev. 2008. PMID: 18929654 Free PMC article. Review. - Parallel Universes for Models of X Chromosome Dosage Compensation in Drosophila: A Review.
Birchler JA. Birchler JA. Cytogenet Genome Res. 2016;148(1):52-67. doi: 10.1159/000445924. Epub 2016 May 12. Cytogenet Genome Res. 2016. PMID: 27166165 Review.
Cited by
- Demasculinization of the Anopheles gambiae X chromosome.
Magnusson K, Lycett GJ, Mendes AM, Lynd A, Papathanos PA, Crisanti A, Windbichler N. Magnusson K, et al. BMC Evol Biol. 2012 May 18;12:69. doi: 10.1186/1471-2148-12-69. BMC Evol Biol. 2012. PMID: 22607633 Free PMC article. - Parallel faster-X evolution of gene expression and protein sequences in Drosophila: beyond differences in expression properties and protein interactions.
Llopart A. Llopart A. PLoS One. 2015 Mar 19;10(3):e0116829. doi: 10.1371/journal.pone.0116829. eCollection 2015. PLoS One. 2015. PMID: 25789611 Free PMC article. - A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence.
Hu TT, Eisen MB, Thornton KR, Andolfatto P. Hu TT, et al. Genome Res. 2013 Jan;23(1):89-98. doi: 10.1101/gr.141689.112. Epub 2012 Aug 30. Genome Res. 2013. PMID: 22936249 Free PMC article. - Non-canonical Drosophila X chromosome dosage compensation and repressive topologically associated domains.
Lee H, Oliver B. Lee H, et al. Epigenetics Chromatin. 2018 Oct 24;11(1):62. doi: 10.1186/s13072-018-0232-y. Epigenetics Chromatin. 2018. PMID: 30355339 Free PMC article. - Non-random genomic integration - an intrinsic property of retrogenes in Drosophila?
Metta M, Schlötterer C. Metta M, et al. BMC Evol Biol. 2010 Apr 28;10:114. doi: 10.1186/1471-2148-10-114. BMC Evol Biol. 2010. PMID: 20426838 Free PMC article.
References
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nature Rev. Genet. 2006;7:645–653. - PubMed
- Hahn MW, Lanzaro GC. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr. Biol. 2005;15:R192–R193. - PubMed
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nature Genet. 2004;36:642–646. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases