Data growth and its impact on the SCOP database: new developments - PubMed (original) (raw)
. 2008 Jan;36(Database issue):D419-25.
doi: 10.1093/nar/gkm993. Epub 2007 Nov 13.
Affiliations
- PMID: 18000004
- PMCID: PMC2238974
- DOI: 10.1093/nar/gkm993
Data growth and its impact on the SCOP database: new developments
Antonina Andreeva et al. Nucleic Acids Res. 2008 Jan.
Abstract
The Structural Classification of Proteins (SCOP) database is a comprehensive ordering of all proteins of known structure, according to their evolutionary and structural relationships. The SCOP hierarchy comprises the following levels: Species, Protein, Family, Superfamily, Fold and Class. While keeping the original classification scheme intact, we have changed the production of SCOP in order to cope with a rapid growth of new structural data and to facilitate the discovery of new protein relationships. We describe ongoing developments and new features implemented in SCOP. A new update protocol supports batch classification of new protein structures by their detected relationships at Family and Superfamily levels in contrast to our previous sequential handling of new structural data by release date. We introduce pre-SCOP, a preview of the SCOP developmental version that enables earlier access to the information on new relationships. We also discuss the impact of worldwide Structural Genomics initiatives, which are producing new protein structures at an increasing rate, on the rates of discovery and growth of protein families and superfamilies. SCOP can be accessed at http://scop.mrc-lmb.cam.ac.uk/scop.
Figures
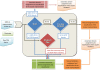
Figure 1.
Workflow of the SCOP update protocol. The update sequence set of new unclassified structures is derived from the PDB SEQRES record. Disordered regions at the termini are masked. The update sequences are clustered using a threshold of 100% identity and 95% coverage for the inclusion of protein sequence into the cluster set. The resulting clusters are used to select a representative sequence set. This dataset is used as a primary input to the pre-classification pipeline. The representative cluster set is first compared using BLAST against itself and a database of non-redundant representative ASTRAL sequences for SCOP domains. This step allows detection of close homologs, usually members of the same SCOP family. Representative sequences without significant sequence match (E-value = 0.001) are further used for two-step PSI-BLAST searches. In the first step, a position-specific scoring matrix (PSSM) is generated by searching the NCBI non-redundant protein database. The resulting PSSM is saved after ten PSI-BLAST iterations or less if the program converges. In the second step, each saved PSSM is used to scan databases of representative ASTRAL and update sequences. In addition, the representative cluster set of unclassified proteins is submitted for RPS-BLAST search against a database of Pfam profiles. The resulting matches are then compared with the matches of pre-computed large-scale comparisons of SCOP domains and Pfam families. A provisional SCOP classification assignment is made for those proteins with a matching region in Pfam that has given a hit to SCOP domain. The results of both RPS-BLAST and PSI-BLAST are used to identify relationships between more distant homologs that are likely to be members of the same SCOP superfamily. Update proteins that are identical or nearly identical to domains classified in the current SCOP release or in the SCOP developmental version are classified automatically. The remaining proteins with and without provisional classification are curated manually.
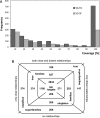
Figure 2.
Statistics of SCOP classification of SG targets. (A) Numbers of SG-families and SG-superfamilies by fraction of SG domains in them. (B) Division of SG-families in ‘true’ and ‘singleton’ families, their SG target contents and their distribution in ‘true’ and ‘singleton’ superfamilies. Note that different parts of the same SG target can be classified into different families and that a ‘true’ superfamily can contain both ‘true’ and ‘singleton’ families.
Similar articles
- SCOP database in 2002: refinements accommodate structural genomics.
Lo Conte L, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Lo Conte L, et al. Nucleic Acids Res. 2002 Jan 1;30(1):264-7. doi: 10.1093/nar/30.1.264. Nucleic Acids Res. 2002. PMID: 11752311 Free PMC article. - SCOP database in 2004: refinements integrate structure and sequence family data.
Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Andreeva A, et al. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D226-9. doi: 10.1093/nar/gkh039. Nucleic Acids Res. 2004. PMID: 14681400 Free PMC article. - The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures.
Andreeva A, Kulesha E, Gough J, Murzin AG. Andreeva A, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D376-D382. doi: 10.1093/nar/gkz1064. Nucleic Acids Res. 2020. PMID: 31724711 Free PMC article. - The SUPERFAMILY database in structural genomics.
Gough J. Gough J. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1897-900. doi: 10.1107/s0907444902015160. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393919 Review. - Structural classification of proteins and structural genomics: new insights into protein folding and evolution.
Andreeva A, Murzin AG. Andreeva A, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Oct 1;66(Pt 10):1190-7. doi: 10.1107/S1744309110007177. Epub 2010 Jul 6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20944210 Free PMC article. Review.
Cited by
- A new definition and properties of the similarity value between two protein structures.
Saberi Fathi SM. Saberi Fathi SM. J Biol Phys. 2016 Oct;42(4):621-636. doi: 10.1007/s10867-016-9429-0. Epub 2016 Sep 13. J Biol Phys. 2016. PMID: 27623799 Free PMC article. - Structure prediction and analysis of DNA transposon and LINE retrotransposon proteins.
Abrusán G, Zhang Y, Szilágyi A. Abrusán G, et al. J Biol Chem. 2013 May 31;288(22):16127-38. doi: 10.1074/jbc.M113.451500. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530042 Free PMC article. - SIFTS: Structure Integration with Function, Taxonomy and Sequences resource.
Velankar S, Dana JM, Jacobsen J, van Ginkel G, Gane PJ, Luo J, Oldfield TJ, O'Donovan C, Martin MJ, Kleywegt GJ. Velankar S, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D483-9. doi: 10.1093/nar/gks1258. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203869 Free PMC article. - Pleiotropic roles of a ribosomal protein in Dictyostelium discoideum.
Amarnath S, Kawli T, Mohanty S, Srinivasan N, Nanjundiah V. Amarnath S, et al. PLoS One. 2012;7(2):e30644. doi: 10.1371/journal.pone.0030644. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363460 Free PMC article. - Molecular modeling comparison of the performance of NS5b polymerase inhibitor (PSI-7977) on prevalent HCV genotypes.
Elfiky AA, Elshemey WM, Gawad WA, Desoky OS. Elfiky AA, et al. Protein J. 2013 Jan;32(1):75-80. doi: 10.1007/s10930-013-9462-9. Protein J. 2013. PMID: 23322006
References
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105192716/MRC_/Medical Research Council/United Kingdom
- G0100305/MRC_/Medical Research Council/United Kingdom
- R01 GM073109/GM/NIGMS NIH HHS/United States
- R01-GM073109/GM/NIGMS NIH HHS/United States
- 077198/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources