RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly - PubMed (original) (raw)
RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly
Michael S Y Huen et al. Cell. 2007.
Abstract
DNA-damage signaling utilizes a multitude of posttranslational modifiers as molecular switches to regulate cell-cycle checkpoints, DNA repair, cellular senescence, and apoptosis. Here we show that RNF8, a FHA/RING domain-containing protein, plays a critical role in the early DNA-damage response. We have solved the X-ray crystal structure of the FHA domain structure at 1.35 A. We have shown that RNF8 facilitates the accumulation of checkpoint mediator proteins BRCA1 and 53BP1 to the damaged chromatin, on one hand through the phospho-dependent FHA domain-mediated binding of RNF8 to MDC1, on the other hand via its role in ubiquitylating H2AX and possibly other substrates at damage sites. Moreover, RNF8-depleted cells displayed a defective G2/M checkpoint and increased IR sensitivity. Together, our study implicates RNF8 as a novel DNA-damage-responsive protein that integrates protein phosphorylation and ubiquitylation signaling and plays a critical role in the cellular response to genotoxic stress.
Figures
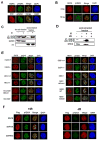
Figure 1. RNF8 is involved in mammalian DNA damage response
A) Localization of tagged RNF8 and Chfr in response to IR. Cells expressing Myc-tagged RNF8 or Chfr were irradiated and immunostained with anti-Myc and anti-pH2AX antibodies. B) Localization of endogenous RNF8 before and after IR treatment in 293T cells. Immunostaining experiments were performed using anti-RNF8 and anti-pH2AX antibodies. C, D) RNF8 relocalizes to chromatin fraction after IR (C), which is reversible following micrococcal nuclease treatment (D). Procedures were carried out as described in Methods and immunoblotting experiments were conducted using indicated antibodies. E) Genetic dependence of RNF8 relocalization following DNA damage. Deficient cells and their respective wild-type counterparts were infected with retrovirus expressing Flag-tagged RNF8. Immunostaining experiments were performed using anti-Flag and anti-γH2AX antibodies. F) The FHA domain, but not the RING domain, of RNF8 targets its localization to DNA damage foci. Cells expressing Flag-tagged wild-type or mutants of RNF8 were mock treated or irradiated and immunostaining were carried out using indicated antibodies.
Figure 2. Structural basis for phosphorylation-dependent binding by RNF8 FHA domain
A) Amino acid selectivity values for the RNF8 FHA domain determined using the phosphothreonine-oriented degenerate peptide library MAXXXX-pT-XXXXAKKK, where X indicates all amino acids except Cys. Values ≥ 1.4 indicate moderate selection; values ≥ 2.0 indicate strong selection. B) Cartoon representation of the RNF8 FHA domain bound to the optimal phosphopeptide ELKpTERY. C) stereo view of the phosphopeptide-binding surface. D) Close up of the phosphate binding pocket, with 2Fo-Fc density map contoured at 2σ. A bound water molecule is evident in the upper center. E–G) Molecular interaction surfaces of the RNF8:phosphopeptide complex, the MDC1 tandem BRCT domain:γ-H2AX phosphopeptide complex, and the Rad53 FHA1:LEVpTEAD phosphopeptide complex. Peptide surfaces are contoured in salmon, protein surfaces are contoured in lime. In the RNF8 FHA domain (E), selection for Tyr over Phe in the +3 position likely results from a water-mediated contact between the Tyr hydroxyl and the backbone nitrogen of Leu-57. In the Rad53 FHA1 structure (G), an Arg residue from the FHA domain occupies the equivalent position as the peptide +3 Tyr in the RNF8 structure (dashed line). (H) Divergence in the phospho-amino acid +3 binding surfaces of the FHA domains of RNF8 and Rad53. The Cα traces of the FHA domains of the RNF8 FHA domain:phosphopeptide complex and the Rad53FHA1 domain:phosphopeptide complex were optimally aligned. The phosphopeptide +3 interacting region is shown in cartoon representation, with the RNF8 FHA domain shaded blue, and its bound phosphopeptide shaded cyan, while the Rad53 FHA1 domain is shaded yellow and its bound phosphopeptide is shaded green. The +3 Tyr residue in the RNF8 optimal phosphopeptide, and the +3 Asp in the Rad53 FHA1 optimal phosphopeptide are shown in stick representation. Note that the +3 Tyr binding site in RNF8 is occluded in the Rad53 FHA domain by an Arg residue that mediates selection for Asp in the +3 position.
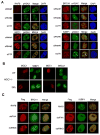
Figure 3. RNF8 is localized to the sites of DNA damage via a FHA-dependent interaction with MDC1
A) Schematic diagram showing full-length MDC1 (WT) and an internal deletion mutant (Del) of MDC1 that abolishes all four putative phosphorylation sites. B) Commassie staining of purified bacterially-expressed GST-RNF8 protein. C) Full length MDC1 but not the deletion mutant (Del) interacts with RNF8 in a pull-down assay. Lysates from 293T cells over-expressing Flag-tagged MDC1 or its deletion mutant were incubated with GST-RNF8 fusion protein immobilized on the glutathione agarose beads for 2 hours before washing and subsequent analysis by Western blotting with anti-Flag antibody. D) MDC1 but not Del mutant of MDC1 co-immunoprecipitates with RNF8. 293T cells were co-transfected with plasmids encoding myc-tagged RNF8 and plasmids encoding SBP-Flag-MDC1 or its deletion mutant. Lysates were incubated with streptavidin beads for 2 hr at 4°C. Thereafter beads were washed three times with NETN, isolates were separated by SDS-PAGE and analyzed by Western blotting using indicated antibodies. E) RNF8 interacts with MDC1 via its FHA domain. Experiments were conducted similar to that described in D) and immunoprecipitation and immunoblotting were carried out as indicated. F) 293T cells were irradiated (10 Gy; 1Gy=100 Rads) or left untreated and cell extract (NETN + 500 mM NaCl) was treated with or without lambda phosphatase prior to diluting and incubating with bacterially expressed 10 μg of GST-RNF8 protein for 2 hr at 4°C. The GST-RNF8 complex was separated by SDS-PAGE to evaluate the amount of endogenous MDC1 that bound specifically to RNF8.
Figure 4. RNF8 is required for accumulation of BRCA1 and 53BP1 at the sites of DNA damage
A) HeLa cells were transfected twice with either RNF8 siRNAs or a non-targeting control siRNA. 48 hr after the second transfection, cells were treated with 10 Gy IR and recovered for 6 hours before they were fixed and permeabilized. Immunostaining experiments were performed as described in the Experimental Procedures. B) 53BP1 and BRCA1 IRIF formation are restored in MDC1 deficient cells reconstituted with full-length MDC1 but not with the deletion mutant of MDC1. Expression constructs encoding HA-tagged MDC1 (WT) or its deletion mutant (Del) were transiently transfected into MDC1 deficient MEFs. 24 hours post-transfection, cells were irradiated (10 Gy) and immuno-stained with indicated antibodies. C) HeLa cells depleted of endogenous RNF8 using siRNA#2 were infected with viruses encoding siRNA-resistant wild-type, delFHA or delRING mutant of RNF8. Infected cells were then irradiated and processed as described above to visualize protein localization as indicated.
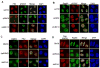
Figure 5. RNF8 functions in concert with UBC13 and is important for IR-induced DNA damage-associated ubiquitin conjugates
A) HeLa cells depleted of endogenous RNF8 or UBC13 were irradiated (10Gy) and immunostained with FK2 and γH2AX antibodies. B) IRIF of UIM-containing protein Rap80 is dependent on RNF8 and UBC13. C) IRIF of damage-associated ubiquitin and D) Rap80 foci formation requires RNF8 FHA and RING domains. HeLa cells infected with virus expressing siRNA-resistant full-length RNF8, delFHA or delRING were transiently transfected with siRNF8#2 to deplete endogenous RNF8. 48 hours after the second transfection, cells were fixed and immuno-stained with indicated antibodies.
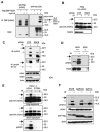
Figure 6. RNF8 is required for H2AX ubiquitylation following DNA damage
A) H2AX is ubiquitylated in vivo. 293T cells were transiently transfected with plasmids encoding myc-tagged ubiquitin with or without plasmids encoding SBP-Flag-H2AX. Immunoprecipitation and immunoblotting were carried out using indicated antibodies. Black arrow indicates doubly ubiquitylated species of H2AX, while grey arrow indicates mono-ubiquitinated H2AX. Multiple-ubiquitinated H2AX species are also pointed out. B) HeLa cells stably expressing HA-tagged H2AX were transfected with control siRNA or RNF8 siRNA were treated with 10 Gy or left untreated. Cells were harvested 1 hr post-irradiation. Cell lysates were prepared, separated by SDS-PAGE and blotted with indicated antibodies. C) HeLa cells transfected with control siRNA or RNF8 siRNA were treated as described (B) and immunoblotting experiments were carried out using indicated antibodies. D) IR-induced H2AX ubiquitylation in H2AX+/+ and H2AX−/− MEFs. Cell lysates prepared from wild-type or H2AX−/− cells before and after irradiation were immunoblotted with anti-H2AX and anti-pH2AX antibodies. E) IR-induced H2AX ubiquitylation requires H2AX phosphorylation. H2AX deficient MEFs stably expressing HA-tagged H2AX or S139A mutant of H2AX were treated with 0 Gy or 10 Gy and immunoblotting was performed using indicated antibodies. F) IR-induced H2AX ubiquitylation requires RNF8 FHA and RING domains. Experiments were carried out as that described in Fig. 5g/5h. Immunoblotting experiments were conducted with antibodies as indicated. Arrow indicates ubiquitylated species of H2AX that only appear after radiation in cells expressing wild-type RNF8.
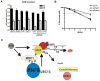
Figure 7. RNF8 is required for G2/M checkpoint control and cell survival following ionizing radiation
A) IR-induced G2/M checkpoint is defective in cells with RNF8 depletion and requires both the RNF8 FHA and RING domains. Summary of the percentages of cells stained positive with phospho-H3 antibody before and after IR treatment from three individual experiments. Error bars indicate standard deviation. HeLa cells were transfected with indicated siRNAs and percentages of mitotic cells before and after radiation were determined by FACS analysis as described in Experimental procedures. B) RNF8-depleted cells display increased radiation sensitivity as determined by colony formation assay. Figure represents value obtained from three separate experiments, each performed in triplicate. Error bars indicate standard deviation. C) A proposed model of the DNA damage responsive pathway involving RNF8. The relocalization of the Rap80-BRCA1 complex and 53BP1 requires RNF8-dependent protein ubiquitylation at the chromatin, whereas the accumulation of NBS1 at DNA damage sites is independent of RNF8.
Similar articles
- RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage.
Sakasai R, Tibbetts R. Sakasai R, et al. J Biol Chem. 2008 May 16;283(20):13549-55. doi: 10.1074/jbc.M710197200. Epub 2008 Mar 12. J Biol Chem. 2008. PMID: 18337245 - RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. Mailand N, et al. Cell. 2007 Nov 30;131(5):887-900. doi: 10.1016/j.cell.2007.09.040. Epub 2007 Nov 20. Cell. 2007. PMID: 18001824 - Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response.
Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Marteijn JA, et al. J Cell Biol. 2009 Sep 21;186(6):835-47. doi: 10.1083/jcb.200902150. J Cell Biol. 2009. PMID: 19797077 Free PMC article. - RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites.
Yan J, Jetten AM. Yan J, et al. Cancer Lett. 2008 Nov 28;271(2):179-90. doi: 10.1016/j.canlet.2008.04.046. Epub 2008 Jun 11. Cancer Lett. 2008. PMID: 18550271 Free PMC article. Review. - Crosstalk between histone modifications during the DNA damage response.
van Attikum H, Gasser SM. van Attikum H, et al. Trends Cell Biol. 2009 May;19(5):207-17. doi: 10.1016/j.tcb.2009.03.001. Epub 2009 Apr 1. Trends Cell Biol. 2009. PMID: 19342239 Review.
Cited by
- RNF4 is required for DNA double-strand break repair in vivo.
Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, Hendriks IA, Radaelli E, Hochepied T, Blanpain C, Sablina A, van Attikum H, Olsen JV, Jochemsen AG, Vertegaal AC, Marine JC. Vyas R, et al. Cell Death Differ. 2013 Mar;20(3):490-502. doi: 10.1038/cdd.2012.145. Epub 2012 Nov 30. Cell Death Differ. 2013. PMID: 23197296 Free PMC article. - A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase.
Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. Gatti M, et al. Cell Cycle. 2012 Jul 1;11(13):2538-44. doi: 10.4161/cc.20919. Epub 2012 Jul 1. Cell Cycle. 2012. PMID: 22713238 Free PMC article. - DNA damage-induced proteasome phosphorylation controls substrate recognition and facilitates DNA repair.
Zhang X, Zhu T, Li X, Zhao H, Lin S, Huang J, Yang B, Guo X. Zhang X, et al. Proc Natl Acad Sci U S A. 2024 Aug 27;121(35):e2321204121. doi: 10.1073/pnas.2321204121. Epub 2024 Aug 22. Proc Natl Acad Sci U S A. 2024. PMID: 39172782 - BRCC3 acts as a prognostic marker in nasopharyngeal carcinoma patients treated with radiotherapy and mediates radiation resistance in vitro.
Tu Z, Xu B, Qu C, Tao Y, Chen C, Hua W, Feng G, Chang H, Liu Z, Li G, Jiang C, Yi W, Zeng M, Xia Y. Tu Z, et al. Radiat Oncol. 2015 May 30;10:123. doi: 10.1186/s13014-015-0427-3. Radiat Oncol. 2015. PMID: 26024915 Free PMC article. - INT6/EIF3E Controls the RNF8-Dependent Ubiquitylation Pathway and Facilitates DNA Double-Strand Break Repair in Human Cells.
Morris C, Tomimatsu N, Burma S, Jalinot P. Morris C, et al. Cancer Res. 2016 Oct 15;76(20):6054-6065. doi: 10.1158/0008-5472.CAN-16-0723. Epub 2016 Aug 22. Cancer Res. 2016. PMID: 27550454 Free PMC article.
References
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. - PubMed
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. - PubMed
- Durocher D, Taylor IADS, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA Domain:phosphopeptide binding specificity and implications for phosphodependent signaling mechanisms. Mol Cell 2000 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA089239-08/CA/NCI NIH HHS/United States
- CA89239/CA/NCI NIH HHS/United States
- R01 CA100109/CA/NCI NIH HHS/United States
- R01 GM060594/GM/NIGMS NIH HHS/United States
- R01 CA089239/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- CA100109/CA/NCI NIH HHS/United States
- CA92312/CA/NCI NIH HHS/United States
- GM 60594/GM/NIGMS NIH HHS/United States
- R01 CA092312-08/CA/NCI NIH HHS/United States
- R01 CA092312/CA/NCI NIH HHS/United States
- R01 CA100109-06/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous