Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis - PubMed (original) (raw)
. 2007 Nov 20;104(47):18682-7.
doi: 10.1073/pnas.0705524104. Epub 2007 Nov 14.
Andrew E L Tee, Antonio Porro, Stewart A Smith, Tanya Dwarte, Pei Yan Liu, Nunzio Iraci, Eric Sekyere, Michelle Haber, Murray D Norris, Daniel Diolaiti, Giuliano Della Valle, Giovanni Perini, Glenn M Marshall
Affiliations
- PMID: 18003922
- PMCID: PMC2141837
- DOI: 10.1073/pnas.0705524104
Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis
Tao Liu et al. Proc Natl Acad Sci U S A. 2007.
Erratum in
- Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):21021
- Correction for Liu et al., Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis.
[No authors listed] [No authors listed] Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2220015120. doi: 10.1073/pnas.2220015120. Epub 2022 Dec 28. Proc Natl Acad Sci U S A. 2023. PMID: 36577078 Free PMC article. No abstract available.
Abstract
Histone deacetylase (HDAC) inhibitors reactivate tumor suppressor gene transcription; induce cancer cell differentiation, growth arrest, and programmed cell death; and are among the most promising new classes of anticancer drugs. Myc oncoproteins can block cell differentiation and promote cell proliferation and malignant transformation, in some cases by modulating target gene transcription. Here, we show that tissue transglutaminase (TG2) was commonly reactivated by HDAC inhibitors in neuroblastoma and breast cancer cells but not normal cells and contributed to HDAC inhibitor-induced growth arrest. TG2 was the gene most significantly repressed by N-Myc in neuroblastoma cells in a cDNA microarray analysis and was commonly repressed by N-Myc in neuroblastoma cells and c-Myc in breast cancer cells. Repression of TG2 expression by N-Myc in neuroblastoma cells was necessary for the inhibitory effect of N-Myc on neuroblastoma cell differentiation. Dual step cross-linking chromatin immunoprecipitation and protein coimmunoprecipitation assays showed that N-Myc acted as a transrepressor by recruiting the HDAC1 protein to an Sp1-binding site in the TG2 core promoter in a manner distinct from it's action as a transactivator at E-Box binding sites. HDAC inhibitor treatment blocked the N-Myc-mediated HDAC1 recruitment and TG2 repression in vitro. In neuroblastoma-bearing N-Myc transgenic mice, HDAC inhibitor treatment induced TG2 expression and demonstrated marked antitumor activity in vivo. Taken together, our data indicate the critical roles of HDAC1 and TG2 in Myc-induced oncogenesis and have significant implications for the use of HDAC inhibitor therapy in Myc-driven oncogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
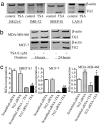
Up-regulation of TG2 by the HDAC inhibitor TSA and its role in cell proliferation in neuroblastoma and breast cancer cells. (a) BE(2)-C, IMR-32, SHEP S1, and LAN-5 neuroblastoma cells were treated with control or 0.1 μM TSA for 6 h, followed by RNA extraction and semiquantitative competitive RT-PCR. (b) MCF-7 and MDA-MB-468 breast cancer cells were treated with control or TSA for 6 or 24 h, followed by RNA extraction and RT-PCR. (c) SHEP S1, MCF-7, and MDA-MB-468 cells were transfected with control or TG2 siRNA for 8 h, followed by treatment with control or TSA for 48 h and incubation with BrDu for the last 6 h. BrDu incorporation was measured as OD units of absorbance. *, P < 0.05 and **, P < 0.01 indicate a statistically significant increase in BrDu incorporation. Error bars indicate standard error.
Fig. 2.
TG2 is transcriptionally repressed by N-Myc and c-Myc in neuroblastoma and breast cancer cells and activated by TSA in nonmalignant cells overexpressing N-Myc or c-Myc. (a) The effect of N-Myc on TG2 gene expression was examined by semiquantitative competitive RT-PCR of mRNA from N-Myc overexpressing SHEP S1 or empty vector control SHEP EV cells. (b) SHEP S1, BE(2)-C, and IMR-32 neuroblastoma cells were transfected with scrambled siRNA or siRNA specifically targeting N-Myc, followed by RNA extraction, and RT-PCR. (c) MCF-7 and MDA-MB-468 breast cancer cells were transfected with scrambled or c-Myc siRNA, followed by RNA extraction and RT-PCR. (d) Nonmalignant MRC-5 and HMEC cells were transfected with a control empty vector or a construct overexpressing c-Myc or N-Myc, respectively, and treated with control or TSA for 24 h. c-Myc, N-Myc, and TG2 expression was examined by RT-PCR.
Fig. 3.
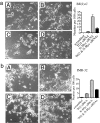
N-Myc blocks neuroblastoma cell differentiation by suppressing TG2 gene transcription. BE(2)-C (a) and IMR-32 (b) cells were transfected with scrambled control siRNA (A), TG2 siRNA (B), N-Myc siRNA (C), or N-Myc siRNA plus TG2 siRNA (D). Five days after transfection, cell differentiation was assessed by analyzing neurite outgrowth under phase contrast microscopy. Cell images were captured and stored, and neurite outgrowth was quantified. Error bars indicate standard error.
Fig. 4.
N-Myc represses TG2 gene transcription by recruiting HDAC1 to the TG2 gene core promoter. (a) Dual cross-linking ChIP and quantitative PCR were applied to LAN-1 cells. Quantitative PCR with primers targeting the Sp1-binding site (Amplicon B) or Amplicon A, 1.6 kb up-stream of TG2 gene transcription start site, was performed in triplicate. (Upper Left) Fold enrichment of a given DNA region immunoprecipitated with anti-N-Myc, Max, Sp1, HDAC1, Tip-60, and TRRAP antibodies was calculated as the ratio between the enrichment obtained with a specific antibody compared with preimmune serum. Results were the average of three independent dual cross-linking ChIP experiments. (Upper Right) Dual cross-linking ChIP was performed on LAN-1 cells treated with control or TSA for 24 h, when a maximal transcriptional reactivation of TG2 was observed. Error bars indicate standard error. Dual ChIP analysis of APEX-1 and nucleolin genes was performed as a control. Amplicons C and E correspond to regions far from the transcription start site. Amplicons D and F, near the transcription start site, carry E-box sequences. (b) Luciferase activity of the two reporters TG2 and ΔTG2, transfected into SHEP TET-OFF cells, was determined in the presence (-TET) or absence (+TET) of N-Myc expression and normalized to that of renilla.
Fig. 5.
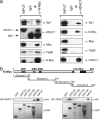
N-Myc directly interacts with Sp1 and HDAC1 through its carboxyl-terminal domain. (a) Protein coimmunoprecipitation (IP) of N-Myc or HDAC1. One milligram of nuclear protein extract from LAN-1 cells was incubated with either a preimmune serum, or an anti-N-Myc antibody (Left) or an anti-HDAC1 antibody (Right). The purified IP-complex was analyzed by Western blot, using antibodies for the following proteins: Sp1, HDAC1, Max, and Tip-60. Lane 1, input; lane 2, preimmune serum IgG IP; lane 3, anti-N-Myc or anti-HDAC1 antibody IP. (b) GST-N-Myc fusion proteins carrying different N-Myc domains were incubated with nuclear extracts expressing HA-HDAC1 or HA-Tip60. GST pull down complexes were analyzed by Western blot analysis, using an anti-HA monoclonal antibody.
Fig. 6.
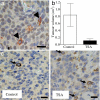
TG2 is expressed exclusively in N-Myc-negative neuroblastoma cells and is up-regulated by an HDAC inhibitor in vivo. (a) Neuroblastoma tissue sections from N-Myc transgenic mice were double-labeled with an anti-N-Myc antibody, which was visualized by using DAB (brown, in the nucleus), and an anti-TG2 antibody, which was visualized by using AEC (red in the cytoplasm, indicated with arrowheads). The nucleus was counterstained with haematoxylin (blue). (b) Neuroblastoma-bearing N-Myc transgenic mice were treated with control or 20 mg per kilogram of body weight of TSA for 1 week before euthanizing. Tumor volume was measured. Error bars indicate standard error. (c) Formalin-fixed tissue sections from the mice were subjected to immunohistochemical assessment of TG2 protein expression, which was visualized by using DAB. The nucleus was counterstained with haematoxylin. Arrows indicate positive cytoplasmic TG2 staining in tumor cells. (Scale bar, 10 μm.)
Similar articles
- Opposing effects of two tissue transglutaminase protein isoforms in neuroblastoma cell differentiation.
Tee AEL, Marshall GM, Liu PY, Xu N, Haber M, Norris MD, Iismaa SE, Liu T. Tee AEL, et al. J Biol Chem. 2010 Feb 5;285(6):3561-3567. doi: 10.1074/jbc.M109.053041. Epub 2009 Dec 10. J Biol Chem. 2010. PMID: 20007697 Free PMC article. - Transcriptional upregulation of histone deacetylase 2 promotes Myc-induced oncogenic effects.
Marshall GM, Gherardi S, Xu N, Neiron Z, Trahair T, Scarlett CJ, Chang DK, Liu PY, Jankowski K, Iraci N, Haber M, Norris MD, Keating J, Sekyere E, Jonquieres G, Stossi F, Katzenellenbogen BS, Biankin AV, Perini G, Liu T. Marshall GM, et al. Oncogene. 2010 Nov 4;29(44):5957-68. doi: 10.1038/onc.2010.332. Epub 2010 Aug 9. Oncogene. 2010. PMID: 20697349 - Tissue transglutaminase (TG2) is involved in the resistance of cancer cells to the histone deacetylase (HDAC) inhibitor vorinostat.
Carbone C, Di Gennaro E, Piro G, Milone MR, Pucci B, Caraglia M, Budillon A. Carbone C, et al. Amino Acids. 2017 Mar;49(3):517-528. doi: 10.1007/s00726-016-2338-5. Epub 2016 Oct 19. Amino Acids. 2017. PMID: 27761756 - Transglutaminase 2 and NF-κB: an odd couple that shapes breast cancer phenotype.
Brown KD. Brown KD. Breast Cancer Res Treat. 2013 Jan;137(2):329-36. doi: 10.1007/s10549-012-2351-7. Epub 2012 Dec 8. Breast Cancer Res Treat. 2013. PMID: 23224146 Review. - Cellular functions of tissue transglutaminase.
Nurminskaya MV, Belkin AM. Nurminskaya MV, et al. Int Rev Cell Mol Biol. 2012;294:1-97. doi: 10.1016/B978-0-12-394305-7.00001-X. Int Rev Cell Mol Biol. 2012. PMID: 22364871 Free PMC article. Review.
Cited by
- The histone deacetylase SIRT2 stabilizes Myc oncoproteins.
Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, Koach J, Tee AE, Haber M, Norris MD, Toon C, Rooman I, Xue C, Cheung BB, Kumar S, Marshall GM, Biankin AV, Liu T. Liu PY, et al. Cell Death Differ. 2013 Mar;20(3):503-14. doi: 10.1038/cdd.2012.147. Epub 2012 Nov 23. Cell Death Differ. 2013. PMID: 23175188 Free PMC article. - Tissue transglutaminase 2 expression is epigenetically regulated in human lung cancer cells and prevents reactive oxygen species-induced apoptosis.
Lee MY, Wu MF, Cherng SH, Chiu LY, Yang TY, Sheu GT. Lee MY, et al. Cancer Manag Res. 2018 Aug 23;10:2835-2848. doi: 10.2147/CMAR.S155582. eCollection 2018. Cancer Manag Res. 2018. PMID: 30197536 Free PMC article. - Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival.
Thole TM, Lodrini M, Fabian J, Wuenschel J, Pfeil S, Hielscher T, Kopp-Schneider A, Heinicke U, Fulda S, Witt O, Eggert A, Fischer M, Deubzer HE. Thole TM, et al. Cell Death Dis. 2017 Mar 2;8(3):e2635. doi: 10.1038/cddis.2017.49. Cell Death Dis. 2017. PMID: 28252645 Free PMC article. - Opposing effects of two tissue transglutaminase protein isoforms in neuroblastoma cell differentiation.
Tee AEL, Marshall GM, Liu PY, Xu N, Haber M, Norris MD, Iismaa SE, Liu T. Tee AEL, et al. J Biol Chem. 2010 Feb 5;285(6):3561-3567. doi: 10.1074/jbc.M109.053041. Epub 2009 Dec 10. J Biol Chem. 2010. PMID: 20007697 Free PMC article. - MYCN-mediated transcriptional repression in neuroblastoma: the other side of the coin.
Gherardi S, Valli E, Erriquez D, Perini G. Gherardi S, et al. Front Oncol. 2013 Mar 11;3:42. doi: 10.3389/fonc.2013.00042. eCollection 2013. Front Oncol. 2013. PMID: 23482921 Free PMC article.
References
- Liu T, Kuljaca S, Tee A, Marshall GM. Cancer Treat Rev. 2006;32:157–165. - PubMed
- Bolden JE, Peart MJ, Johnstone RW. Nat Rev Drug Discov. 2006;5:769–784. - PubMed
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Nat Rev Cancer. 2001;1:194–202. - PubMed
- Kelly WK, Marks PA. Nat Clin Pract Oncol. 2005;2:150–157. - PubMed
- Johnstone RW. Nat Rev Drug Discov. 2002;1:287–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous