Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster - PubMed (original) (raw)
Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster
Marta L Wayne et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Sexual selection drives faster evolution in males. The X chromosome is potentially an important target for sexual selection, because hemizygosity in males permits accumulation of alleles, causing tradeoffs in fitness between sexes. Hemizygosity of the X could cause fundamentally different modes of inheritance between the sexes, with more additive variation in males and more nonadditive variation in females. Indeed, we find that genetic variation for the transcriptome is primarily additive in males but nonadditive in females. As expected, these differences are more pronounced on the X chromosome than the autosomes, but autosomal loci are also affected, possibly because of X-linked transcription factors. These differences may be of evolutionary significance because additive variation responds quickly to selection, whereas nonadditive genetic variation does not. Thus, hemizygosity of the X may underlie much of the faster male evolution of the transcriptome and potentially other phenotypes. Consistent with this prediction, genes that are additive in males and nonadditive in females are overrepresented among genes responding to selection for increased mating speed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
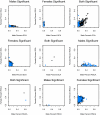
Fig. 1.
Percent variance explained for different components of genetic variation, plotted by sex. Blue squares represent genes on the X; black circles represent genes on the autosomes. Genes could be either significant for the particular term in one sex (males or females) or significant in both sexes (both significant). Plots are not presented for genes significant in SCA for males, for RGCA for females, or for RSCA for both sexes, because there were no significant genes in any of these categories.
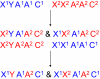
Fig. 2.
A reciprocal cross: In the first cross, parent 1 (blue) is the sire, and parent 2 (red) is the dam; in the second, parent 2 (red) is the sire and parent 1 (blue) is the dam. Sons differ between pairs of reciprocal crosses for their single X chromosome, but daughters from both crosses are heterozygous for the X. Within each reciprocal cross, sons and daughters have identical cytoplasms but different X genotypes because sons are hemizygous, whereas daughters are heterozygous diploids. Reciprocal crosses also differ in both males and females for epigenetic and parent-of-origin effects, not illustrated here.
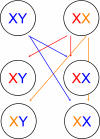
Fig. 3.
Each X chromosome from a female has an equal probability of being transmitted to a son or a daughter [red and orange Xs; the X from males is always transmitted to daughters, never to sons (blue X and arrow)].
Similar articles
- Autosomal and X-Linked Additive Genetic Variation for Lifespan and Aging: Comparisons Within and Between the Sexes in Drosophila melanogaster.
Griffin RM, Schielzeth H, Friberg U. Griffin RM, et al. G3 (Bethesda). 2016 Dec 7;6(12):3903-3911. doi: 10.1534/g3.116.028308. G3 (Bethesda). 2016. PMID: 27678519 Free PMC article. - Reduced X-linked rare polymorphism in males in comparison to females of Drosophila melanogaster.
Takahashi KH, Tanaka K, Itoh M, Takano-Shimizu T. Takahashi KH, et al. J Hered. 2009 Jan-Feb;100(1):97-105. doi: 10.1093/jhered/esn078. Epub 2008 Oct 3. J Hered. 2009. PMID: 18836147 - The X chromosome is a hot spot for sexually antagonistic fitness variation.
Gibson JR, Chippindale AK, Rice WR. Gibson JR, et al. Proc Biol Sci. 2002 Mar 7;269(1490):499-505. doi: 10.1098/rspb.2001.1863. Proc Biol Sci. 2002. PMID: 11886642 Free PMC article. - Transcriptional modulation of entire chromosomes: dosage compensation.
Lucchesi JC. Lucchesi JC. J Genet. 2018 Jun;97(2):357-364. J Genet. 2018. PMID: 29932054 Review. - Pathology from evolutionary conflict, with a theory of X chromosome versus autosome conflict over sexually antagonistic traits.
Frank SA, Crespi BJ. Frank SA, et al. Proc Natl Acad Sci U S A. 2011 Jun 28;108 Suppl 2(Suppl 2):10886-93. doi: 10.1073/pnas.1100921108. Epub 2011 Jun 20. Proc Natl Acad Sci U S A. 2011. PMID: 21690397 Free PMC article. Review.
Cited by
- Genome-Wide Association Study of Circadian Behavior in Drosophila melanogaster.
Harbison ST, Kumar S, Huang W, McCoy LJ, Smith KR, Mackay TFC. Harbison ST, et al. Behav Genet. 2019 Jan;49(1):60-82. doi: 10.1007/s10519-018-9932-0. Epub 2018 Oct 19. Behav Genet. 2019. PMID: 30341464 Free PMC article. - The Y Chromosome Modulates Splicing and Sex-Biased Intron Retention Rates in Drosophila.
Wang M, Branco AT, Lemos B. Wang M, et al. Genetics. 2018 Mar;208(3):1057-1067. doi: 10.1534/genetics.117.300637. Epub 2017 Dec 20. Genetics. 2018. PMID: 29263027 Free PMC article. - What the X has to do with it: differences in regulatory variability between the sexes in Drosophila simulans.
Graze RM, McIntyre LM, Morse AM, Boyd BM, Nuzhdin SV, Wayne ML. Graze RM, et al. Genome Biol Evol. 2014 Apr;6(4):818-29. doi: 10.1093/gbe/evu060. Genome Biol Evol. 2014. PMID: 24696400 Free PMC article. - Genome-wide gene expression effects of sex chromosome imprinting in Drosophila.
Lemos B, Branco AT, Jiang PP, Hartl DL, Meiklejohn CD. Lemos B, et al. G3 (Bethesda). 2014 Jan 10;4(1):1-10. doi: 10.1534/g3.113.008029. G3 (Bethesda). 2014. PMID: 24318925 Free PMC article. - Identification of Genes Contributing to a Long Circadian Period in Drosophila Melanogaster.
Kumar S, Tunc I, Tansey TR, Pirooznia M, Harbison ST. Kumar S, et al. J Biol Rhythms. 2021 Jun;36(3):239-253. doi: 10.1177/0748730420975946. Epub 2020 Dec 4. J Biol Rhythms. 2021. PMID: 33274675 Free PMC article.
References
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Science. 2002;297:2270–2275. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials