Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition - PubMed (original) (raw)
Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition
Mohammad M Mohammad et al. EMBO J. 2007.
Abstract
The Tetrahymena thermophila ribosomal DNA (rDNA) replicon contains dispersed cis-acting replication determinants, including reiterated type I elements that associate with sequence-specific, single-stranded binding factors, TIF1 through TIF4. Here, we show that TIF4, previously implicated in cell cycle-controlled DNA replication and rDNA gene amplification, is the T. thermophila origin recognition complex (TtORC). We further demonstrate that TtORC contains an integral RNA subunit that participates in rDNA origin recognition. Remarkably, this RNA, designated 26T, spans the terminal 282 nts of 26S ribosomal RNA. 26T RNA exhibits extensive complementarity to the type I element T-rich strand and binds the rDNA origin in vivo. Mutations that disrupt predicted interactions between 26T RNA and its complementary rDNA target change the in vitro binding specificity of ORC and diminish in vivo rDNA origin utilization. These findings reveal a role for ribosomal RNA in chromosome biology and define a new mechanism for targeting ORC to replication initiation sites.
Figures
Figure 1
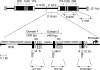
Schematic of the T. thermophila rDNA minichromosome. ‘Palindromic' rDNA encodes the 17S, 5.8S and 26S rRNAs. Domains 1 and 2 (D1, D2) are an imperfect duplication and contain coordinately regulated origins (Ori). Reiterated type I elements (IA–ID), pause site elements (PSE1–3) and type III elements are shown. RFB, developmentally regulated replication fork barrier; ovals, positioned nucleosomes. DNA segments subjected to PCR amplification in Figures 3 and 6 are shown (D1 Ori, D2 Ori, promoter, 26S rRNA gene middle and 3′ end fragments).
Figure 2
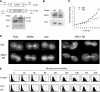
Functional analysis of ORC1 knockdown mutants. (A) Disruption of the ORC1 gene in the polyploid macronucleus. Top: schematic of wild-type ORC1 and the ORC1 disruption allele (H3: _Hin_DIII sites). Bottom: Southern blot analysis (wild type: 3.8 kb; disruption: 5.6 kb). WT: strain CU428; KD: ORC1 knockdown strain, TD101. TIF1 served as a control. (B) Northern analysis of WT and ORC1 KD strains. (C) Growth curves of WT (TX615) and ORC1 knockdown (TD101) strains. TX615 encodes an MTT1-NEO transgene targeted to the MTT1 locus, and was grown in the same pm dosage as TD101 (1 mg/ml). (D) Nuclear division in control (WT, TX615) and ORC1 knockdown (KD) cells visualized with acridine orange. Left panel: the stages of cytokinesis were defined by the extent of constriction of the cytokinetic furrow. Right panel: additional examples of aberrant macronuclear divisions. (E) Flow cytometry of control (TX615) and ORC1 knockdown (TD101) strains synchronized by starvation and re-feeding.
Figure 3
In vitro and in vivo analysis of TIF4/ORC. (A) Western blot analysis of TAP-tagged Orc1p. Strains: WT, CU428; TAP-tagged ORC1, TD102; TAP/APT (TAP-tagged ORC1 and aptamer-tagged 26T RNA), TD152. Ponceau S staining of nuclear extracts served as a loading control. (B) ChIP analysis of TAP-tagged ORC complexes. PCR of input and immunoprecipitated DNA from crosslinked chromatin preparations without [(−) Ab] or with [(+) Ab] antibody against the Orc1p TAP tag. Data from two representative independent ChIP experiments are shown. A 2.5- to 5-fold enrichment for the Domain 1 rDNA origin fragment was observed. See Figure 1 for PCR primer locations. (C) Standard EMSA analysis with wild-type nuclear extracts (strain CU428) and labeled type I element oligos (C3 rDNA type IB element T-rich strand, T51: 5′ CTCAAAAGTTGCAAAAGTTCGGAAGGTTTACTATT TTTGTTTTTTTTTTT; A-rich strand, A53: 5′ GGCAAAAAAAAAAACAAAAATAGTAAACCTTCCGA ACTTTTGCAACTTTTGAG) or T51:A53 duplex (DS). (D) Western blot EMSA of Orc1p and Orc2p. Nuclear extracts were prepared from wild-type (CU428) and TAP-tagged ORC1 (TD102) strains. DNA substrates were not labeled. Following native gel electrophoresis and transfer, membranes were probed with Orc1p or Orc2p antibodies to monitor their migration. Lanes MN, RN and DN: MNase-, RNase A- and DNase I-digested extracts.
Figure 4
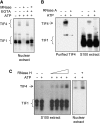
Tetrahymena ORC is a ribonucleoprotein complex. (A) MNase-sensitive DNA binding. Nuclear extracts were sequentially incubated with MNase and EGTA prior to EMSA with the type I element T strand substrate, C3-ssT51. Unbound DNA was run off the gel to better resolve TIF4/ORC–DNA complexes. In the absence of EGTA, gel shift complexes were not observed due to degradation of substrate DNA. (B) RNase A-sensitive binding to the rDNA type IB element T-rich strand. Oligo affinity-purified ORC or S100 extracts were treated with RNase A prior to EMSA analysis. (C) Enhanced binding to the type I element T-rich strand following RNase H treatment. Gel shift complexes were formed on ice and digested with RNase H (S100 extract: 1–5 U; nuclear extract: 5 U) prior to EMSA.
Figure 5
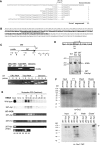
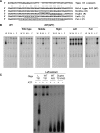
Characterization of the ORC-associated RNA. (A) Sequence of partial cDNAs derived from oligonucleotide affinity-purified ORC. Top line: DNA sequence of a 26S RNA-coding region segment (sense strand). Remaining sequences: 5′ end of sequenced cDNAs. The dashed region contains 26 nt of complete sequence identity. (B) Complete sequence of 26T RNA starting at the 5′ end of the longest cloned cDNA. The boldfaced segment spans the 26S rRNA D12 expansion region. Underlined residues correspond to the binding site for oligo-dT-directed reverse transcription during cDNA cloning. (C) Characterization of 26T RNA. Upper panel: schematic of the 26S rRNA-coding region and primers used for RT–PCR analysis (Ter: 3′ end of mature 26S rRNA). Middle panel: RT–PCR test for contaminating full-length 26S rRNA. Primers A and B amplify the middle segment of 26S rRNA. Substrates: affinity-purified ORC RNA, total cellular RNA and genomic DNA. The 5′ residue of forward (A) and reverse (B) primers map to nt 5369 and 5624 (EMBL accession number M11155). Lower panel: 3′ end mapping of 26T RNA. RT was performed with primers 1, 2, 3 or 4 (5′ nt positions: 8472, 8534, 8608 and 8756, respectively). cDNAs were amplified following addition of primer 5 (5′ nt position: 8343). Substrates were as noted above. (D) Southern blot analysis of 26T-Ext and 26T-Apt transformants. Upper panel: schematic of transformation constructs. TER: telomerase RNA gene 5′ and 3′ flanking segments. Lower panel: Southern blot analysis of _Eco_RI-digested DNA from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) transformant strains (probe: 26T DNA fragment). (E) Incorporation of transgenic 26T RNAs into an RNP complex. Nuclear extracts from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) strains were fractionated on a 24 ml Superdex 200 fast protein liquid chromatography column (GE Healthcare). Fractions (0.3 ml) were assayed for ORC binding to the type I element T-rich strand (ssT51) and 26T RNA. Migration of molecular weight markers are depicted with asterisks (left to right: 665, 430, 230 and 153 kD). The migration of free 26T-Apt RNA was determined by pretreating extracts with proteinase K (RT–PCR, lower panel). (F) Orc1p and Orc2p associate with aptamer-tagged 26T RNA. Nuclear extracts from wild-type (CU428), 26T-Ext (MM201) and 26T-Apt (MM202) strains were incubated with streptavidin (SA) sepharose. Orc2p western blot analysis was performed on bound and unbound fractions. FT: flow through/unbound fraction. For Orc1p analysis, TD151 and TD152 were used. Both strains produce TAP-tagged Orc1p. TD152 (T) encodes 26T-Ext RNA, while TD151 (A) encodes SA-binding 26T-Apt RNA. Arrows: full-length Orc1p and Orc2p. Prior to pull-down analysis, nuclear extracts from the different strains were normalized for protein concentration. Here, 10% of each sample (input, flow through, wash and eluate) was subjected to western blot analysis.
Figure 6
Sequence alignment and in vitro targeting of 26T RNA to the rDNA origin. (A) Alignment of 26T RNA and the type I element T-rich strand. Underlined DNA residues correspond to sequences present in all four type I elements (IA–ID). (B) Chromatin association of 26T RNA and Orc2p. RT–PCR (26T-Apt RNA) and western blot analysis of mock and DNase I-treated nuclei following centrifugation to separate insoluble (pellet) and soluble (supernatant) fractions (strain MM202). (C) Representative streptavidin chromatin pull-down experiment. Formaldehyde crosslinked chromatin from 26T-Ext (MM201) and 26T-Apt transformants (MM202: C1; MM203: C2) was subjected to PCR amplification following purification on SA sepharose beads. The 5′ end position of forward (F) and reverse complementary (R) PCR primers are designated in their name: origin (F744, R1075), promoter (F1580, R1887), 26S middle (F5369, R5624), 26S 3′ end (F8343, R8472) (see Figure 1 schematic). A minimum three-fold enrichment of the rDNA origin region was observed in four independent experiments with 26T-Apt transformant strains.
Figure 7
Gel shift analysis of 26T RNA base-pairing mutants. (A) Sequence of wild-type (WT) and 26T RNA derivatives (M, R, L and F) carrying mutations in the predicted type I element T-strand base-pairing region. (B) EMSA analysis of nuclear extracts from wild-type and 26T RNA mutants with the wild-type (W) substrate, C3ssT51, and derivatives (M, R, L and F) that re-establish complementary with the respective mutant RNA. (C) Further characterization of the 26T-Apt(L) mutant.
Figure 8
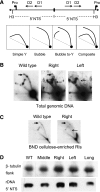
Analysis of rDNA replication intermediates in 26T RNA base-pairing mutants. (A) Schematic of the palindromic rDNA 5′ NTS fragment generated by _Hin_DIII digestion, and possible RI patterns following 2D gel electrophoresis. Simple Y arc: passive replication of 5′ NTS origins; bubble or bubble-to-Y arcs: initiation within the 5′ NTS; composite (simple Y arc and bubbles): active and passive replication of the 5′ NTS. (B) Southern blot analysis of _Hin_DIII-digested DNA resolved on a neutral–neutral 2D gel. Thick right-pointing arrows point to bubble arcs. Thin left-pointing arrows point to simple Y arcs. (C) 2D gel analysis following enrichment for RIs on BND cellulose. (D) Southern blot analysis of total genomic DNA from 26T RNA transformants.
Similar articles
- Differential targeting of Tetrahymena ORC to ribosomal DNA and non-rDNA replication origins.
Donti TR, Datta S, Sandoval PY, Kapler GM. Donti TR, et al. EMBO J. 2009 Feb 4;28(3):223-33. doi: 10.1038/emboj.2008.282. Epub 2009 Jan 15. EMBO J. 2009. PMID: 19153611 Free PMC article. - Characterization of a novel origin recognition complex-like complex: implications for DNA recognition, cell cycle control, and locus-specific gene amplification.
Mohammad M, York RD, Hommel J, Kapler GM. Mohammad M, et al. Mol Cell Biol. 2003 Jul;23(14):5005-17. doi: 10.1128/MCB.23.14.5005-5017.2003. Mol Cell Biol. 2003. PMID: 12832485 Free PMC article. - Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome.
Kapler GM. Kapler GM. Curr Opin Genet Dev. 1993 Oct;3(5):730-5. doi: 10.1016/s0959-437x(05)80091-7. Curr Opin Genet Dev. 1993. PMID: 8274855 Review. - The ribosomal RNA genes of Tetrahymena: structure and function.
Engberg J. Engberg J. Eur J Cell Biol. 1985 Jan;36(1):133-51. Eur J Cell Biol. 1985. PMID: 3884336 Review.
Cited by
- Developmentally Programmed Switches in DNA Replication: Gene Amplification and Genome-Wide Endoreplication in Tetrahymena.
Meng X, Dang HQ, Kapler GM. Meng X, et al. Microorganisms. 2023 Feb 16;11(2):491. doi: 10.3390/microorganisms11020491. Microorganisms. 2023. PMID: 36838456 Free PMC article. - Transcriptome analysis of the binucleate ciliate Tetrahymena thermophila with asynchronous nuclear cell cycles.
Zhang L, Cervantes MD, Pan S, Lindsley J, Dabney A, Kapler GM. Zhang L, et al. Mol Biol Cell. 2023 Feb 1;34(2):rs1. doi: 10.1091/mbc.E22-08-0326. Epub 2022 Dec 7. Mol Biol Cell. 2023. PMID: 36475712 Free PMC article. - An Optimized and Versatile Counter-Flow Centrifugal Elutriation Workflow to Obtain Synchronized Eukaryotic Cells.
Liu Y, Nan B, Niu J, Kapler GM, Gao S. Liu Y, et al. Front Cell Dev Biol. 2021 Apr 20;9:664418. doi: 10.3389/fcell.2021.664418. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33959616 Free PMC article. - Time-course profiling of bovine alphaherpesvirus 1.1 transcriptome using multiplatform sequencing.
Moldován N, Torma G, Gulyás G, Hornyák Á, Zádori Z, Jefferson VA, Csabai Z, Boldogkői M, Tombácz D, Meyer F, Boldogkői Z. Moldován N, et al. Sci Rep. 2020 Nov 24;10(1):20496. doi: 10.1038/s41598-020-77520-1. Sci Rep. 2020. PMID: 33235226 Free PMC article. - The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses.
Sheng Y, Duan L, Cheng T, Qiao Y, Stover NA, Gao S. Sheng Y, et al. Sci China Life Sci. 2020 Oct;63(10):1534-1542. doi: 10.1007/s11427-020-1689-4. Epub 2020 Apr 13. Sci China Life Sci. 2020. PMID: 32297047 Free PMC article.
References
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 42: 833–837 - PubMed
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA-replication by a multiprotein complex. Nature 357: 128–134 - PubMed
- Bielinsky AK, Gerbi SA (1998) Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279: 95–98 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources