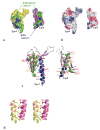The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus - PubMed (original) (raw)
Comparative Study
The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus
Marissa E Yanez et al. J Mol Biol. 2008.
Abstract
Type II secretion systems (T2SS) translocate virulence factors from the periplasmic space of many pathogenic bacteria into the extracellular environment. The T2SS of Vibrio cholerae and related species is called the extracellular protein secretion (Eps) system that consists of a core of multiple copies of 11 different proteins. The pseudopilins, EpsG, EpsH, EpsI, EpsJ and EpsK, are five T2SS proteins that are thought to assemble into a pseudopilus, which is assumed to interact with the outer membrane pore, and may actively participate in the export of proteins. We report here biochemical evidence that the minor pseudopilins EpsI and EpsJ from Vibrio species interact directly with one another. Moreover, the 2.3 A resolution crystal structure of a complex of EspI and EpsJ from Vibrio vulnificus represents the first atomic resolution structure of a complex of two different pseudopilin components from the T2SS. Both EpsI and EpsJ appear to be structural extremes within the family of type 4a pilin structures solved to date, with EpsI having the smallest, and EpsJ the largest, "variable pilin segment" seen thus far. A high degree of sequence conservation in the EpsI:EpsJ interface indicates that this heterodimer occurs in the T2SS of a large number of bacteria. The arrangement of EpsI and EpsJ in the heterodimer would correspond to a right-handed helical character of proteins assembled into a pseudopilus.
Figures
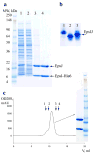
Figure 1. The interaction between soluble domains of EpsI and EpsJ from V. cholerae
(a) An SDS-PAGE gel of a non-tagged soluble domain of _Vc_EpsJ with the first 24 residues truncated co-eluting with a His-tagged soluble domain of similarly truncated _Vc_EpsI. Lysate from three liters of cell culture expressing truncated _Vc_EpsJ were mixed with lysate from one liter of cell culture expressing _Vc_EpsI-His6. The mixed cell lysate (Lane 1) was then applied to a column containing 1 ml of Ni-NTA resin. The flowthrough (Lane 2) and a subsequent 10 ml wash (Lane 3) with 20 mM imidazole were collected. Bound protein was eluted (Lane 4) with 200 mM imidazole. Molecular weight markers are indicated on the left. (b) A native PAGE gel of _Vc_EpsI (Lane 1), _Vc_EpsJ (Lane 2) and EpsI-EpsJ complex (Lane3). Approximately 2 mgs of purified protein was loaded per lane. (c) A Size Exclusion Chromatography profile of the _Vc_EpsI-EpsJ complex on a Superdex75 column. The positions of molecular weight standards are indicated by arrows: 1 - bovine serum albumin (67 kDa), 2 - chicken ovalbumin (43 kDa), 3 -myoglobin (17.6 kDa) and 4 - ribonuclease A (13.7 kDa). The insert represents an SDS-PAGE gel of the peak fraction showing that the soluble domains of _Vc_EpsI and _Vc_EpsJ maintain an approximately stoichiometric complex during purification. The same molecular weight markers are used as in (a).
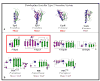
Figure 2. EpsI/GspI and EpsJ/GspJ family sequence alignment
A Multalin alignment of _Vv_EpsI and _Vv_EpsJ with nine EpsI/GspI and EpsJ/GspJ homologues. A black triangle denotes the start of the constructs for _Vv_EpsI and _Vv_EpsJ used for crystallization. The secondary structure elements as identified by DSSP are shown on top, with the conserved N-terminal α-helix in blue, the variable segment in purple and the conserved β-sheet depicted in green. Invariant residues are highlighted with a red background, while residues that are conserved in all but 1 or 2 homologues are highlighted in pink. Residues that are a hydrophobic residue in all 10 homologues are highlighted orange. Yellow background denotes medium sequence conservation and white background denotes poor sequence conservation. In the lower two lines, contact residues that form Van-der-Waals contacts in the EpsI-EpsJ interface are represented by triangles and residues that form intermolecular H-bonds are denoted by stars. Contact residues in the EpsI:EpsJ interface are labeled with triangles and stars in blue, those in the EpsI:EpsJ’ interface in green, and those in the EpsJ:EpsJ’ interface in red. The solvent accessibility, as assigned by ESPRIPT on the basis of the _Vv_EpsI:EpsJ structure, is denoted by the bar acc, with dark blue meaning residues that are highly accessible, cyan meaning residues that are partially accessible and white denoting residues that are buried.
Figure 3. The crystal structure of the V. vulnificus EpsI:EpsJ complex
(a) Ribbon diagram of one _Vv_EpsI2:EpsJ2 heterotetramer. _Vv_EpsI colored in blue, _Vv_EpsI’ in cyan, _Vv_EpsJ in brown and _Vv_EpsJ’ in red. One chloride ion, located between _Vv_EpsJ and _Vv_EpsJ’, is shown in green. (b) A “top” view of one _Vv_EpsI2:EpsJ2 heterotetramer with buried accessible surfaces (in Å2) of the intersubunit contacts indicated. (c) A close-up view of one _Vv_EpsI:EpsJ heterodimer, in the same orientation as the heterotetramer shown in (A) above, with _Vv_EpsI colored in dark blue and _Vv_EpsJ colored in brown. (d) The _Vv_EpsI:EpsJ heterodimer shown in (C) rotated by 180° with each of the secondary structure elements in _Vv_EpsI and _Vv_EpsJ labeled. Two small stretches of residues in _Vv_EpsJ (67-71 and 132 - 133) that could not be included in the model due to disorder, are denoted by a dotted black line.
Figure 4. Comparison of V. vulnificus EpsI and EpsJ soluble domains with known Type 4a-like Pilin structures
(a) The minor pseudopilins _Vv_EpsI and _Vv_EpsJ are shown schematically along side the equivalent structures of a minor pseudopilin EpsH from V. cholerae (Yanez et. al., submitted) and of the major pseudopilin GspG from K. oxytoca (PDB code 1T92). To facilitate comparison, all structures are shown with the same orientation of the conserved β-sheet colored in green. (b) Topology diagrams of the globular domains of four T2SS pseudopilins with known structure (V. vulnificus EpsI, V. vulnificus EpsJ, V. cholerae EpsM, K. oxytoca GspG) and of the three T4BP pilins PilAPAK from P. aeruginosa (PDB code 1DZO), PilE from N. gonorrhoeae (PDB code 2HI2), and PilAK1224 from P. aeruginosa (PDB code 1RGO). The red box contains the new structures of _Vv_EpsI and _Vv_EpsJ. Both these minor pseudopilins exhibit an “atypical fold” compared to known Type 4a Pilin structures as described in the text.
Figure 5. The V. vulnificus EpsI:EpsJ Interface
_Vv_EpsI and _Vv_EpsJ are shown in the same “butterfly” orientation in all figures (a)-(e). (a) The contact surface of the _Vv_EpsI:EpsJ interface. N-terminally truncated _Vv_EpsJ in yellow and N-terminally truncated _Vv_EpsI in blue. All residues that form hydrophobic contacts between _Vv_EpsJ and _Vv_EpsI are shown in green, while residues that form polar contacts between _Vv_EpsJ and _Vv_EpsI are colored blue. (b) An electrostatic representation of the _Vv_EpsI:EpsJ interface. The I:J interface is largely hydrophobic, with a small negatively charged region in _Vv_EpsI that contacts a small positively charged surface in _Vv_EpsJ. (c) A cartoon representation of the _Vv_EpsI:EpsJ interface showing the sequence conservation of the contact residues in _Vv_EpsI and _Vv_EpsJ. The cartoon representation of _Vv_EpsJ is colored according to the Type 4a Pilin structural elements: the N-terminal helix in blue, the variable segment green and the C-terminal β-sheet purple. The cartoon representation of _Vv_EpsI is colored in grey. The contact residues are denoted by spheres. Contact residues in _Vv_EpsJ are labeled with spheres that are colored according to the Type 4a Pilin structural elements. Contact residues in _Vv_EpsI colored according to the _Vv_EpsJ residues with which they interact. Contact residues are labeled according to their sequence conservation within the EpsI/GspI and EpsJ/GspJ families as follows: invariant residues by a red box; hydrophobic residues labeled with a red residue name and number; residues with moderate sequence conservation labeled in yellow, and residues with low sequence conservation labeled in black. One residue, _Vv_EpsI-Asp45, a conserved negatively charged interface residue, is labeled in pink. (d) The “helix-helix” interactions between _Vv_EpsI and _Vv_EpsJ. The helix of _Vv_EpsJ colored in yellow, the helix of _Vv_EpsI in purple. Six residues from the helix of _Vv_EpsJ form hydrophobic interactions with six residues along the helix of _Vv_EpsI. The residues, shown in stick representation, are labeled according to their sequence conservation as in (c) above.
Figure 6. The shape and sequence conservation of the EpsI:EpsJ heterodimer
(a) Stereo view of the _Vv_EpsI:EpsJ heterodimer shown in the same orientation as in figure 3. _Vv_EpsI and _Vv_EpsJ are colored by element and per chain: oxygens and nitrogens in red and blue, respectively; carbons in _Vv_EpsI in green; and carbons in _Vv_EpsJ in yellow. Conserved residues facing the viewer in the deep crevice are Asp45, Arg106, Tyr108 of EpsI, and Gln45 and Gln57 of EpsJ. In addition, Leu 48 and Leu 189 of EpsJ are consistently hydrophobic in the EpsJ/GspJ family (Fig. 2). (b) The heterodimer in (a) above is rotated about the x-axis by 90° to show the “top” view of one _Vv_EpsI:EpsJ heterodimer. This representation shows the large crevice formed between the two pseudopilins _Vv_EpsI and _Vv_EpsJ.
Figure 7. The _Vv_EpsI:EpsJ heterodimer corresponds with a right handed one-start helical pseudopilin assembly
(a) A view of the _Vv_EpsI:EpsJ heterodimer approximately perpendicular to the two N-terminal helices of _Vv_EpsI and _Vv_EpsJ with the N-termini of the helices at the lower part of the figure. In the crystal structure the N-terminal helix of _Vv_EpsI is shifted “upwards” by about 12 Å with respect to _Vv_EpsJ. These helices are thought to run approximately parallel to the pseudopilus fiber axis assuming that the T2SS pseudopilus shares this feature with the pilus model of Craig et al. 2006. _Vv_EpsJ is depicted in brown, _Vv_EpsI colored in blue. (b) View of the _Vv_EpsI:EpsJ heterodimer approximately parallel to the axes of the N-terminal helices, approximately 90 degrees rotated with respect to (A) above, with the N-termini of the helices in front. Since the helix axes are probably aligned roughly parallel to the pseudopilus helix axis, the symmetry operation relating EpsJ and EpsI corresponds with a right-handed rotation as indicated by the arrow.
Similar articles
- Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
Lam AY, Pardon E, Korotkov KV, Hol WGJ, Steyaert J. Lam AY, et al. J Struct Biol. 2009 Apr;166(1):8-15. doi: 10.1016/j.jsb.2008.11.008. Epub 2008 Dec 10. J Struct Biol. 2009. PMID: 19118632 Free PMC article. - Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae.
Yanez ME, Korotkov KV, Abendroth J, Hol WG. Yanez ME, et al. J Mol Biol. 2008 Mar 14;377(1):91-103. doi: 10.1016/j.jmb.2007.08.041. Epub 2007 Aug 23. J Mol Biol. 2008. PMID: 18241884 Free PMC article. - The 1.59Å resolution structure of the minor pseudopilin EpsH of Vibrio cholerae reveals a long flexible loop.
Raghunathan K, Vago FS, Grindem D, Ball T, Wedemeyer WJ, Bagdasarian M, Arvidson DN. Raghunathan K, et al. Biochim Biophys Acta. 2014 Feb;1844(2):406-15. doi: 10.1016/j.bbapap.2013.11.013. Epub 2013 Dec 4. Biochim Biophys Acta. 2014. PMID: 24316251 - Architecture, Function, and Substrates of the Type II Secretion System.
Korotkov KV, Sandkvist M. Korotkov KV, et al. EcoSal Plus. 2019 Feb;8(2):10.1128/ecosalplus.ESP-0034-2018. doi: 10.1128/ecosalplus.ESP-0034-2018. EcoSal Plus. 2019. PMID: 30767847 Free PMC article. Review. - Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion.
Hansen JK, Forest KT. Hansen JK, et al. J Mol Microbiol Biotechnol. 2006;11(3-5):192-207. doi: 10.1159/000094054. J Mol Microbiol Biotechnol. 2006. PMID: 16983195 Review.
Cited by
- Improvement of the crystallizability and expression of an RNA crystallization chaperone.
Ravindran PP, Héroux A, Ye JD. Ravindran PP, et al. J Biochem. 2011 Nov;150(5):535-43. doi: 10.1093/jb/mvr093. Epub 2011 Jul 23. J Biochem. 2011. PMID: 21785128 Free PMC article. - On the path to uncover the bacterial type II secretion system.
Douzi B, Filloux A, Voulhoux R. Douzi B, et al. Philos Trans R Soc Lond B Biol Sci. 2012 Apr 19;367(1592):1059-72. doi: 10.1098/rstb.2011.0204. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22411978 Free PMC article. Review. - The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae.
Abendroth J, Mitchell DD, Korotkov KV, Johnson TL, Kreger A, Sandkvist M, Hol WG. Abendroth J, et al. J Struct Biol. 2009 Jun;166(3):303-15. doi: 10.1016/j.jsb.2009.03.009. Epub 2009 Mar 24. J Struct Biol. 2009. PMID: 19324092 Free PMC article. - Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
Zhang Y, Faucher F, Zhang W, Wang S, Neville N, Poole K, Zheng J, Jia Z. Zhang Y, et al. PLoS Pathog. 2018 Oct 22;14(10):e1007343. doi: 10.1371/journal.ppat.1007343. eCollection 2018 Oct. PLoS Pathog. 2018. PMID: 30346996 Free PMC article. - Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
Lam AY, Pardon E, Korotkov KV, Hol WGJ, Steyaert J. Lam AY, et al. J Struct Biol. 2009 Apr;166(1):8-15. doi: 10.1016/j.jsb.2008.11.008. Epub 2008 Dec 10. J Struct Biol. 2009. PMID: 19118632 Free PMC article.
References
- Gerritse G, Ure R, Bizoullier F, Quax WJ. The phenotype enhancement method identifies the Xcp outer membrane secretion machinery from Pseudomonas alcaligenes as a bottleneck for lipase production. J Biotechnol. 1998;64:23–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI034501/AI/NIAID NIH HHS/United States
- R01 AI034501-07/AI/NIAID NIH HHS/United States
- R56 AI034501/AI/NIAID NIH HHS/United States
- AI34501/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources