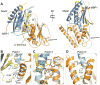p31comet blocks Mad2 activation through structural mimicry - PubMed (original) (raw)
p31comet blocks Mad2 activation through structural mimicry
Maojun Yang et al. Cell. 2007.
Abstract
The status of spindle checkpoint signaling depends on the balance of two opposing dynamic processes that regulate the highly unusual two-state behavior of Mad2. In mitosis, a Mad1-Mad2 core complex recruits cytosolic Mad2 to kinetochores through Mad2 dimerization and converts Mad2 to a conformer amenable to Cdc20 binding, thereby facilitating checkpoint activation. p31(comet) inactivates the checkpoint through binding to Mad1- or Cdc20-bound Mad2, thereby preventing Mad2 activation and promoting the dissociation of the Mad2-Cdc20 complex. Here, we report the crystal structure of the Mad2-p31(comet) complex. The C-terminal region of Mad2 that undergoes rearrangement in different Mad2 conformers is a major structural determinant for p31(comet) binding, explaining the specificity of p31(comet) toward Mad1- or Cdc20-bound Mad2. p31(comet) adopts a fold strikingly similar to that of Mad2 and binds at the dimerization interface of Mad2. Thus, p31(comet) exploits the two-state behavior of Mad2 to block its activation by acting as an "anti-Mad2."
Figures
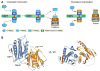
Figure 1. Structure of the Mad2-p31comet Complex
(A) Schematic drawing of the proposed mechanisms of Mad2 activation by the Mad1-Mad2 core complex and the inhibition of this process by p31comet. Upon checkpoint activation, autoinhibited O-Mad2 binds to the Mad1-Mad2 core complex through Mad2-Mad2 dimerization, which induces a conformational change of O-Mad2 and converts it into an activated intermediate state (I-Mad2). I-Mad2 dissociates from the Mad1-Mad2 core complex to become the active conformer, C-Mad2, with or without Cdc20. During checkpoint inactivation, p31comet binds to the Mad1-Mad2 core complex and blocks the binding of O-Mad2, thus preventing the generation of I-Mad2 and C-Mad2. p31comet also binds to Cdc20-bound Mad2 and activates APC/C. The symbols used for different Mad2 conformers are shown in the shaded yellow box. The Mad2-binding motif of Mad1 is colored red. (B) Ribbon diagram of the Mad2-p31comet complex in two views. Mad2, p31comet, and MBP1 are colored blue, orange, and red, respectively. The N- and C-termini of Mad2 and p31comet are labeled. All structural figures were generated with PyMOL (
).
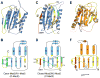
Figure 2. p31comet Has a Fold Similar to Mad2
Ribbon and topology diagrams of O-Mad2 (PDB ID 1DUJ) (A,B), C-Mad2 (C,D) and p31comet (E,F). The secondary structural elements are labeled and the missing loops in C-Mad2 (residues 108-118) and p31comet (residues 97-118) are shown as dashed lines. The core domain is colored blue for Mad2 and orange for p31comet. The N- and C-terminal regions involved in Mad2 conformational change are colored yellow, except that residues 172-175 and 184-192 are in green. The analogous segment in p31comet is shown in yellow. MBP1 and the C-terminal pseudo-ligand tail of p31comet are shown in red.
Figure 3. Mad2 and p31comet Share Limited Sequence Similarity
(A) Structure-based sequence alignment of human Mad2 and p31comet. The secondary structural elements of Mad2 are drawn above the sequences and colored blue. The secondary structural elements of p31comet are drawn below the sequences and colored orange. MBP1 is colored red. R35 and E98 in Mad2 are aligned with R84 and E163 in p31comet respectively, and are labeled with asterisks. The YQRXXΦP motif is boxed. In the alignment, Mad2 and p31comet share less than 10% sequence identity. (B) The buried salt bridge between R35 and E98 in Mad2. (C) The buried salt bridge between R84 and E163 in p31comet.
Figure 4. Interactions between Mad2 and p31comet
(A) Ribbon diagrams of the Mad2-p31comet complex. Two different views are shown to provide a clearer perspective of the Mad2-p31comet interface. Helix αC in Mad2 is colored cyan to highlight its central role in establishing interactions between Mad2 and p31comet. Three main patches of interactions at the Mad2-p31comet interface are labeled and circled with red dashed lines. (B-D) Interactions between Mad2 and p31comet. The side chains of contacting residues are shown as sticks. Nitrogen and oxygen atoms are colored blue and red, respectively. Mad2 carbons are colored yellow while p31comet carbons are colored gray and labeled in italics. The tightly bound water molecules are drawn as red spheres in (C).
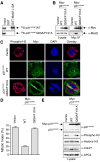
Figure 5. Mad2 Binding Is Required for the Function of p31comet in Checkpoint Silencing
(A) 35S-labeled p31comet WT or Q83A/F191A proteins were incubated with beads bound to GST or GST-Mad2 L13A. After washing, proteins bound to beads were separated on SDS-PAGE and analyzed using a phosphoimager. (B) HeLa cells were transfected with the indicated plasmids and treated with nocodazole. Lysates of the transfected cells were immunoprecipitated using anti-Myc beads. Both the lysates and immunoprecipitates were blotted with the indicated antibodies. (C) HeLa cells were transfected with the indicated plasmids, treated with nocodazole, and stained with DAPI (blue), anti-Myc (green), and anti-phospho-H3 (red). Scale bars indicate 10 μm. (D) The mitotic indices of the transfected cells in (C) were quantified. At least 400 cells were counted for each transfection. The averages and standard deviations of three separate experiments are shown. (E) Lysates of cells described in (C-D) were blotted with the indicated antibodies.
Figure 6. A Structure Model for the Blockage of Mad1-assisted Mad2 Activation by p31comet
(A) A structural model of the Mad1-Mad2-p31comet complex. The Mad2 molecule in the Mad2-p31comet complex was superimposed with the Mad2 molecules in the Mad1-Mad2 complex (PDB ID 1GO4). For clarity, the Mad2 monomers in the Mad1-Mad2 complex are omitted. Mad1 is colored green with its Mad2-binding region colored red. The three interacting helices in Mad2-p31comet are indicated. (B) A surface representation to show that C-Mad2 uses a similar surface for the binding of p31comet or O-Mad2. The p31comet-binding residues of C-Mad2 are colored yellow and the four key interacting residues, R133, Q134, R184 and F141, are colored red. The O-Mad2-binding residues of C-Mad2 are colored yellow. The same four residues R133, Q134, R184 and F141 (red) that are important for p31comet binding are also involved in O-Mad2 binding. (C) Ribbon diagram of the O-Mad2–C-Mad2 dimer (Mapelli et al., 2007). O-Mad2 is colored in cyan. C-Mad2 is colored blue with its C-terminal region shown in yellow. MBP1 is in red. The αC helices are labeled. (D) Ribbon diagram of the Mad2-p31comet complex with C-Mad2 in the same orientation as in (C). (E) Overlay of ribbon diagrams of the O-Mad2–C-Mad2 dimer and the Mad2-p31comet complex. The C-Mad2 molecules in both structures are superimposed. The color scheme is the same as in (C) and (D).
Similar articles
- p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit.
Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. Westhorpe FG, et al. J Cell Sci. 2011 Nov 15;124(Pt 22):3905-16. doi: 10.1242/jcs.093286. Epub 2011 Nov 18. J Cell Sci. 2011. PMID: 22100920 Free PMC article. - Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint.
Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Xia G, et al. EMBO J. 2004 Aug 4;23(15):3133-43. doi: 10.1038/sj.emboj.7600322. Epub 2004 Jul 15. EMBO J. 2004. PMID: 15257285 Free PMC article. - Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer.
Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. Yang M, et al. PLoS Biol. 2008 Mar 4;6(3):e50. doi: 10.1371/journal.pbio.0060050. PLoS Biol. 2008. PMID: 18318601 Free PMC article. - Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model.
Yu H. Yu H. J Cell Biol. 2006 Apr 24;173(2):153-7. doi: 10.1083/jcb.200601172. J Cell Biol. 2006. PMID: 16636141 Free PMC article. Review. - The Mad2-Binding Protein p31comet as a Potential Target for Human Cancer Therapy.
Henriques AC, Silva PMA, Sarmento B, Bousbaa H. Henriques AC, et al. Curr Cancer Drug Targets. 2021;21(5):401-415. doi: 10.2174/1568009621666210129095726. Curr Cancer Drug Targets. 2021. PMID: 33511944 Review.
Cited by
- Chromosome Division in Early Embryos-Is Everything under Control? And Is the Cell Size Important?
Horakova A, Konecna M, Anger M. Horakova A, et al. Int J Mol Sci. 2024 Feb 9;25(4):2101. doi: 10.3390/ijms25042101. Int J Mol Sci. 2024. PMID: 38396778 Free PMC article. Review. - A role of WT1 in cell division and genomic stability.
Shandilya J, Roberts SG. Shandilya J, et al. Cell Cycle. 2015;14(9):1358-64. doi: 10.1080/15384101.2015.1021525. Cell Cycle. 2015. PMID: 25789599 Free PMC article. Review. - CAMK2D serves as a molecular scaffold for RNF8-MAD2 complex to induce mitotic checkpoint in glioma.
Chuah YH, Tay EXY, Grinchuk OV, Yoon J, Feng J, Kannan S, Robert M, Jakhar R, Liang Y, Lee BWL, Wang LC, Lim YT, Zhao T, Sobota RM, Lu G, Low BC, Crasta KC, Verma CS, Lin Z, Ong DST. Chuah YH, et al. Cell Death Differ. 2023 Aug;30(8):1973-1987. doi: 10.1038/s41418-023-01192-3. Epub 2023 Jul 19. Cell Death Differ. 2023. PMID: 37468549 Free PMC article. - Targeting TRIP13 for overcoming anticancer drug resistance (Review).
Zhao L, Ye S, Jing S, Gao YJ, He T. Zhao L, et al. Oncol Rep. 2023 Nov;50(5):202. doi: 10.3892/or.2023.8639. Epub 2023 Oct 6. Oncol Rep. 2023. PMID: 37800638 Free PMC article. - Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Brulotte ML, Jeong BC, Li F, Li B, Yu EB, Wu Q, Brautigam CA, Yu H, Luo X. Brulotte ML, et al. Nat Commun. 2017 Dec 5;8(1):1956. doi: 10.1038/s41467-017-02012-2. Nat Commun. 2017. PMID: 29208896 Free PMC article.
References
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. - PubMed
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. - PubMed
- Consortium TC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. - PubMed
- Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D Biol Crystallogr. 1998;54:487–493. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases