Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR - PubMed (original) (raw)
Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR
Ioannis Gelis et al. Cell. 2007.
Abstract
Recognition of signal sequences by cognate receptors controls the entry of virtually all proteins to export pathways. Despite its importance, this process remains poorly understood. Here, we present the solution structure of a signal peptide bound to SecA, the 204 kDa ATPase motor of the Sec translocase. Upon encounter, the signal peptide forms an alpha-helix that inserts into a flexible and elongated groove in SecA. The mode of binding is bimodal, with both hydrophobic and electrostatic interactions mediating recognition. The same groove is used by SecA to recognize a diverse set of signal sequences. Impairment of the signal-peptide binding to SecA results in significant translocation defects. The C-terminal tail of SecA occludes the groove and inhibits signal-peptide binding, but autoinhibition is relieved by the SecB chaperone. Finally, it is shown that SecA interconverts between two conformations in solution, suggesting a simple mechanism for polypeptide translocation.
Figures
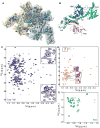
Figure 1. Methyl-TROSY spectra of the full-length 204-kDa SecA
(A) Structural model of dimeric E. coli SecA (PDB 2FSF) displayed as semi-transparent solvent-accessible surface. The methyl groups are displayed as spheres. The color code is as follows; Ile, orange (54 residues); Leu, light blue (82 residues); Val, dark blue (59 residues); Met, green (33 residues). The two protomers are colored differently. (B) Structural model of one of the protomers of dimeric E. coli SecA colored according to domain organization. (C-E) 1H-13C HMQC spectra of SecA (C) U-[2H,12C],Val,Leu-[13CH3,12CD3], (D) U-[2H,12C],Ile-δ1-[13CH3], and (E) U-[2H,12C],Met-[13CH3]. In (C), expanded views of the crowded areas of the spectrum are shown. In (D), regions of the spectrum of unliganded SecA (orange) are overlaid with the spectrum of SecA bound to the KRR-LamB signal sequence (magenta).
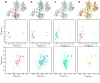
Figure 2. Strategy for the assignment of methyl correlations of SecA
Each column in the figure displays a structural model of one of the protomers of SecA with the domain or fragment studied in isolation being highlighted, along with the corresponding 1H-13C HMQC of Ile-δ1 methyls (displayed as spheres in the model) and the backbone 1H-15N HSQC. (A) PBD (residues 220-379). (B) SecAΔC/ΔIRA2 (residues 1-420, comprising NBD and PBD). (C) SecAΔC (residues 1-610, comprising NBD, PBD and IRA2). (D) Full-length SecA (residues 1-901). Only few resonances for the backbone of the full-length SecA are visible (Figure S9).
Figure 3. Structural basis for signal peptide recognition by SecA
(A) The lowest-energy structure of SecA bound to the KRR-LamB signal peptide is shown. SecA is displayed as a semi-transparent solvent-accessible surface and the signal peptide is shown in yellow. A ribbon model is displayed below the surface (color code is as in Figure 1B). (B) Closer view of the groove bound to the signal peptide. Green and red surface indicates hydrophobic and acidic residues, respectively. Peptide is shown as a ribbon ball-and-stick representation and most of its residues are numbered. (C) Contacts between the peptide (shown in yellow) and SecA residues. Electrostatic and hydrophobic interactions are indicated with red and green lines, respectively. SecA residues are colored according to the domain they are located at. (D) A view of the groove bound to the signal peptide, wherein SecA is shown in ribbons. The peptide orientation is similar to that in (C). Dotted lines indicate electrostatic interactions between basic peptide residues and acidic SecA residues. Primed numbers indicate peptide residues.
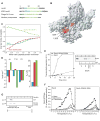
Figure 4. Signal peptide binding to SecA and the effect of its impairment on the function of SecA
(A) The four different signal sequences used in the present study are shown. In KRR-LamB, the asterisks indicate the positions where a single Cys mutation was introduced, followed by the incorporation of the nitroxide spin label. Basic and hydrophobic residues are colored blue and green, respectively. (B) Chemical shift mapping of the interaction of the signal peptides shown in (A) with SecA. The magenta colored region indicates the common residues whose chemical shift is significantly affected upon signal peptide binding (see text for details). (C) Binding isotherms of the calorimetric titration of the KRR-LamB signal peptide to SecA (open, cyan circles), SecA-I304A/L306A (filled, red circles), and SecA in 160 mM K+ buffer (green squares). In the first two cases, SecA is in 40 mM K+ buffer. Ile304 and Leu306, whose position within the groove is indicated in (B), were mutated to assess the relative contribution of hydrophobic interactions in SecA-signal peptide binding, whereas higher buffer salt was used to assess the contribution of electrostatic interactions. (D) Thermodynamic parameters of the calorimetric titrations in (C) displayed as bars. The color code is as in (C). Weakening of the electrostatic interactions results in hydrophobic interactions being the dominant driving force for complex formation, as suggested by the observation that the reaction becomes enthalpically unfavorable, but strongly entropically favorable. (E) ATP-driven in vitro translocation of proOmpA-His into SecYEG-containing IMVs catalyzed by SecA or SecA-I304A/L306A (right panel). Lane 1: 5% of undigested proOmpA-His input. Lane 5: membranes were dissolved with Triton x-100 (1%) prior to proteinase K addition. Proteins were TCA-precipitated, analyzed by SDS-PAGE and immunostained with α-His antibody. Left panel: Time-course of proOmpA translocation kinetics. 20 A.U. corresponds to 1.6 pmol of translocated proOmpA. (F) kcat values (pmoles Pi/pmol SecA protomer per min) of basal, membrane and translocation ATPase activities of SecA as a function of temperature. Averaged data of three repetitions are shown. (G) IMV flotation assay showing weaker binding of proOmpA to SecYEG-bound SecA- I304A/L306A than to wild-type SecA.
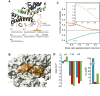
Figure 5. Inhibition of signal peptide binding by the C-tail of SecA
(A) View of the C-tail of B. subtilis SecA (PDB 1M6N). This is the only crystal structure wherein part of the C-tail is resolved. The E. coli SecA sequence of the C-tail is shown below the model. Dotted lines indicate crystallographically unresolved regions. Red lines indicate the zinc-finger region, which is the primary SecB binding site. (B) Structural modeling of the interaction of the C-tail in E. coli SecA. The C-tail is shown in orange surface and it partially occludes the peptide binding groove. (C) Binding isotherms of the calorimetric titration of the wild-type LamB signal peptide to SecA (open, cyan circles), SecA834 (filled, red circles), and SecA bound to SecB (green squares). (D) Thermodynamic parameters of the calorimetric titrations in (C) displayed as bars. The color code is as in (C).
Figure 6. SecA interconverts between an open and closed conformation in solution
(A) SecA shown in the so-called open (left) and closed (right) conformations. Interconversion between the two conformations requires that PBD undergo a ~60° rigid-body rotation (Osborne et al., 2004). PBD is displayed as semi-transparent surface. The green sphere indicates residue 830, where a paramagnetic spin label was introduced. Residues Ile304 and Ile789 are shown as yellow and red spheres, respectively. Characteristic distances in the two conformations are indicated. A strong NOE between Ile304 and Ile789 was observed demonstrating that SecA adopts predominantly the open conformation in solution. (B) Overlaid 1H-13C HMQC spectra of SecA bearing a spin label in position 830 in the reduced (blue) and oxidized (red) state. Residues that approach the spin label, even transiently, experience a broadening effect, which is suppressed in the reduced state.
Figure 7. Model of the SecA-mediated preprotein translocation
PBD is shown in the closed state in E and in the open state in all other panels. The C-tail is not shown in D-F for clarity. See text for details.
Similar articles
- The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA.
Chou YT, Gierasch LM. Chou YT, et al. J Biol Chem. 2005 Sep 23;280(38):32753-60. doi: 10.1074/jbc.M507532200. Epub 2005 Jul 26. J Biol Chem. 2005. PMID: 16046390 - Characterization of the Escherichia coli SecA signal peptide-binding site.
Grady LM, Michtavy J, Oliver DB. Grady LM, et al. J Bacteriol. 2012 Jan;194(2):307-16. doi: 10.1128/JB.06150-11. Epub 2011 Nov 4. J Bacteriol. 2012. PMID: 22056930 Free PMC article. - NMR structure of the C-terminal domain of SecA in the free state.
Matousek WM, Alexandrescu AT. Matousek WM, et al. Biochim Biophys Acta. 2004 Nov 1;1702(2):163-71. doi: 10.1016/j.bbapap.2004.08.012. Biochim Biophys Acta. 2004. PMID: 15488768 - The structural view of bacterial translocation-specific chaperone SecB: implications for function.
Zhou J, Xu Z. Zhou J, et al. Mol Microbiol. 2005 Oct;58(2):349-57. doi: 10.1111/j.1365-2958.2005.04842.x. Mol Microbiol. 2005. PMID: 16194224 Review. - Structure and function of SecA, the preprotein translocase nanomotor.
Vrontou E, Economou A. Vrontou E, et al. Biochim Biophys Acta. 2004 Nov 11;1694(1-3):67-80. doi: 10.1016/j.bbamcr.2004.06.003. Biochim Biophys Acta. 2004. PMID: 15546658 Review.
Cited by
- Hold the fold: how delayed folding aids protein secretion.
McCaul N, Braakman I. McCaul N, et al. EMBO J. 2022 Dec 1;41(23):e112787. doi: 10.15252/embj.2022112787. Epub 2022 Oct 31. EMBO J. 2022. PMID: 36314692 Free PMC article. - How Quality Control Systems AID Sec-Dependent Protein Translocation.
Jiang C, Wynne M, Huber D. Jiang C, et al. Front Mol Biosci. 2021 Apr 13;8:669376. doi: 10.3389/fmolb.2021.669376. eCollection 2021. Front Mol Biosci. 2021. PMID: 33928127 Free PMC article. Review. - Defining the Escherichia coli SecA dimer interface residues through in vivo site-specific photo-cross-linking.
Yu D, Wowor AJ, Cole JL, Kendall DA. Yu D, et al. J Bacteriol. 2013 Jun;195(12):2817-25. doi: 10.1128/JB.02269-12. Epub 2013 Apr 12. J Bacteriol. 2013. PMID: 23585536 Free PMC article. - N-terminal domain of human Hsp90 triggers binding to the cochaperone p23.
Karagöz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rüdiger SG. Karagöz GE, et al. Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):580-5. doi: 10.1073/pnas.1011867108. Epub 2010 Dec 23. Proc Natl Acad Sci U S A. 2011. PMID: 21183720 Free PMC article. - A 3D time-shared NOESY experiment designed to provide optimal resolution for accurate assignment of NMR distance restraints in large proteins.
Mishra SH, Harden BJ, Frueh DP. Mishra SH, et al. J Biomol NMR. 2014 Dec;60(4):265-74. doi: 10.1007/s10858-014-9873-8. Epub 2014 Nov 9. J Biomol NMR. 2014. PMID: 25381567 Free PMC article.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. - PubMed
- Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. - PubMed
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. - PubMed
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Bio Crystallogr. 1998;54:905–921. - PubMed
- Chou YT, Gierasch LM. The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA. J Biol Chem. 2005;280:32753–32760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases