Virus movement maintains local virus population diversity - PubMed (original) (raw)
Comparative Study
. 2007 Nov 27;104(48):19102-7.
doi: 10.1073/pnas.0709445104. Epub 2007 Nov 19.
Affiliations
- PMID: 18025457
- PMCID: PMC2141915
- DOI: 10.1073/pnas.0709445104
Comparative Study
Virus movement maintains local virus population diversity
Jamie C Snyder et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Viruses are the largest reservoir of genetic material on the planet, yet little is known about the population dynamics of any virus within its natural environment. Over a 2-year period, we monitored the diversity of two archaeal viruses found in hot springs within Yellowstone National Park (YNP). Both temporal phylogeny and neutral biodiversity models reveal that virus diversity in these local environments is not being maintained by mutation but rather by high rates of immigration from a globally distributed metacommunity. These results indicate that geographically isolated hot springs are readily able to exchange viruses. The importance of virus movement is supported by the detection of virus particles in air samples collected over YNP hot springs and by their detection in metacommunity sequencing projects conducted in the Sargasso Sea. Rapid rates of virus movement are not expected to be unique to these archaeal viruses but rather a common feature among virus metacommunities. The finding that virus immigration rather than mutation can dominate community structure has significant implications for understanding virus circulation and the role that viruses play in ecology and evolution by providing a reservoir of mobile genetic material.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
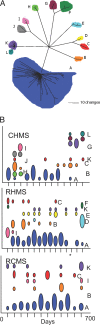
Fig. 1.
General water chemistry of each hot spring monitor site. Although not entirely static, the overall geochemical signature of each spring was distinct and relatively stable over the 2-year sampling course. Pie charts illustrate the distinct geochemical signature of each hot spring. (A) CHMS. (B) RCMS. (C) RHMS. Average values are reported in the table.
Fig. 2.
Comparison of the archaeal community detected at each of the hot springs. (A) CHMS. (B) RHMS. (C) RCMS. Pie charts illustrate the average relative abundance of each genus detected by using 16S rRNA gene analysis. The community composition was relatively stable over the 2-year time course, with Sulfolobus being the only common, but not the dominant, resident in each of the three hot springs. A, Acidianus; C, Caldococcus; D, Desulfurococcus; M, Metallosphaera; P, Pyrodictium; T, Thermocladium; V, Vulcanisaeta; S, Stygiolobus; O, other.
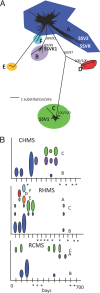
Fig. 3.
Phylogenetic relationships of clades of SIRV and their detection through time. (A) SIRV phylogenetic tree composed of 2,165 clones from three hot springs over all sampling times. Twelve well supported clades (A–L) with both maximum parsimony bootstrap values (10,000 resampling) and maximum likelihood clade credibility values are indicated. (B) Dominant SIRV clades detected over time within CHMS, RHMS, and RCMS. The size of the oval represents the relative number of sequences belonging to a particular clade detected at each sampling time. Asterisks indicate sampling times in which SIRV sequences were not detected.
Fig. 4.
A diverse SSV community transiently visits geochemically distinct hot spring environments. (A) SSV phylogenetic tree composed of 779 clones from three hot springs from all sampling times shows six well supported clades (A–F), with both maximum parsimony bootstrap values (10,000 resamplings) and maximum likelihood clade credibility values indicated. Globally distributed SSV isolates (indicated by isolate abbreviation) cluster within YNP clades. (B) Dominant SSV clades detected over time within the three hot springs, where the size of the oval represents the relative number of sequences for each clade. Asterisks indicate sampling times in which SSV sequences were not detected.
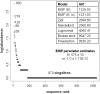
Fig. 5.
Rank abundance distribution of 2,944 sequences in 1,042 sequence types for SIRV and SSV sampled at all three hot springs during the ≈2-year study. Distributions and their AIC values are shown, with EtSF being the best fit. The abundances of the five most abundant genotypes are reported next to the corresponding point.
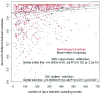
Fig. 6.
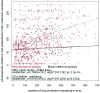
Community composition of SIRV and SSV as a function of time. Pairwise comparisons of community composition (Jaccard's distance) for SIRV (open circles) and SSV (bullets) are shown. The models including within-hot-springs (black lower lines) and among-hot-springs (upper red lines) comparisons along with days between sampling are illustrated for SIRV and SSV. The center lines depict the model, including only days between sampling. The 95% confidence intervals for the mantel correlation of each of the two models were essentially identical.
Fig. 7.
Phylogenetic relatedness of SIRV and SSV as a function of time. The phylogenetic distance (net-relatedness index) between samples compared with days between sampling are shown for both SIRV (open circles) and SSV (filled circles). The upper (red) and lower (black) lines represent the models, including both days between sampling events and among-hot-springs or within-hot-springs comparisons. The model including only days between sampling events is shown by the center line. The 95% confidence intervals for the mantel correlation of each of the two models were essentially identical.
Similar articles
- Differentiation and Structure in Sulfolobus islandicus Rod-Shaped Virus Populations.
Bautista MA, Black JA, Youngblut ND, Whitaker RJ. Bautista MA, et al. Viruses. 2017 May 19;9(5):120. doi: 10.3390/v9050120. Viruses. 2017. PMID: 28534836 Free PMC article. - Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs.
Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. Bolduc B, et al. J Virol. 2012 May;86(10):5562-73. doi: 10.1128/JVI.07196-11. Epub 2012 Feb 29. J Virol. 2012. PMID: 22379100 Free PMC article. - New archaeal viruses discovered by metagenomic analysis of viral communities in enrichment cultures.
Liu Y, Brandt D, Ishino S, Ishino Y, Koonin EV, Kalinowski J, Krupovic M, Prangishvili D. Liu Y, et al. Environ Microbiol. 2019 Jun;21(6):2002-2014. doi: 10.1111/1462-2920.14479. Epub 2019 Jan 8. Environ Microbiol. 2019. PMID: 30451355 Free PMC article. - Hot crenarchaeal viruses reveal deep evolutionary connections.
Ortmann AC, Wiedenheft B, Douglas T, Young M. Ortmann AC, et al. Nat Rev Microbiol. 2006 Jul;4(7):520-8. doi: 10.1038/nrmicro1444. Epub 2006 Jun 6. Nat Rev Microbiol. 2006. PMID: 16755285 Review. - Advances in understanding archaea-virus interactions in controlled and natural environments.
Snyder JC, Young MJ. Snyder JC, et al. Curr Opin Microbiol. 2011 Aug;14(4):497-503. doi: 10.1016/j.mib.2011.07.007. Epub 2011 Aug 5. Curr Opin Microbiol. 2011. PMID: 21821465 Review.
Cited by
- A global atlas of soil viruses reveals unexplored biodiversity and potential biogeochemical impacts.
Graham EB, Camargo AP, Wu R, Neches RY, Nolan M, Paez-Espino D, Kyrpides NC, Jansson JK, McDermott JE, Hofmockel KS; Soil Virosphere Consortium. Graham EB, et al. Nat Microbiol. 2024 Jul;9(7):1873-1883. doi: 10.1038/s41564-024-01686-x. Epub 2024 Jun 20. Nat Microbiol. 2024. PMID: 38902374 Free PMC article. - Viruses of the Turriviridae: an emerging model system for studying archaeal virus-host interactions.
Overton MS, Manuel RD, Lawrence CM, Snyder JC. Overton MS, et al. Front Microbiol. 2023 Sep 21;14:1258997. doi: 10.3389/fmicb.2023.1258997. eCollection 2023. Front Microbiol. 2023. PMID: 37808280 Free PMC article. Review. - Metavirome and its functional diversity analysis through microbiome study of the Sikkim Himalayan hot spring solfataric mud sediments.
Das S, Kumari A, Sherpa MT, Najar IN, Thakur N. Das S, et al. Curr Res Microb Sci. 2020 May 30;1:18-29. doi: 10.1016/j.crmicr.2020.05.002. eCollection 2020 Sep. Curr Res Microb Sci. 2020. PMID: 34841298 Free PMC article. - Unveiling Ecological and Genetic Novelty within Lytic and Lysogenic Viral Communities of Hot Spring Phototrophic Microbial Mats.
Guajardo-Leiva S, Santos F, Salgado O, Regeard C, Quillet L, Díez B. Guajardo-Leiva S, et al. Microbiol Spectr. 2021 Dec 22;9(3):e0069421. doi: 10.1128/Spectrum.00694-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787442 Free PMC article. - T4-like myovirus community shaped by dispersal and deterministic processes in the South China Sea.
Li H, Liu L, Wang Y, Cai L, He M, Wang L, Hu C, Jiao N, Zhang R. Li H, et al. Environ Microbiol. 2021 Feb;23(2):1038-1052. doi: 10.1111/1462-2920.15290. Epub 2020 Nov 3. Environ Microbiol. 2021. PMID: 33089595 Free PMC article.
References
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, et al. Nature. 2005;437:1162–1166. - PubMed
- Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Science. 2002;296:1439–1443. - PubMed
- de Oliveira T, Pybus O, Rambaut A, Salemi M, Cassol S, Cicozzi M, Rezza G, Gattinara GC, D'Arrigo R, Amicosante M, et al. Nature. 2006;444:836–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources