PRIDE: new developments and new datasets - PubMed (original) (raw)
. 2008 Jan;36(Database issue):D878-83.
doi: 10.1093/nar/gkm1021. Epub 2007 Nov 22.
Affiliations
- PMID: 18033805
- PMCID: PMC2238846
- DOI: 10.1093/nar/gkm1021
PRIDE: new developments and new datasets
Philip Jones et al. Nucleic Acids Res. 2008 Jan.
Abstract
The PRIDE (http://www.ebi.ac.uk/pride) database of protein and peptide identifications was previously described in the NAR Database Special Edition in 2006. Since this publication, the volume of public data in the PRIDE relational database has increased by more than an order of magnitude. Several significant public datasets have been added, including identifications and processed mass spectra generated by the HUPO Brain Proteome Project and the HUPO Liver Proteome Project. The PRIDE software development team has made several significant changes and additions to the user interface and tool set associated with PRIDE. The focus of these changes has been to facilitate the submission process and to improve the mechanisms by which PRIDE can be queried. The PRIDE team has developed a Microsoft Excel workbook that allows the required data to be collated in a series of relatively simple spreadsheets, with automatic generation of PRIDE XML at the end of the process. The ability to query PRIDE has been augmented by the addition of a BioMart interface allowing complex queries to be constructed. Collaboration with groups outside the EBI has been fruitful in extending PRIDE, including an approach to encode iTRAQ quantitative data in PRIDE XML.
Figures
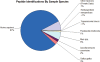
Figure 1.
PRIDE includes data for 25 different species. This pie chart illustrates the representation of each species in terms of the total number of peptide identifications for each species.
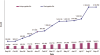
Figure 2.
This graph illustrates the increasing redundancy of the peptide identifications submitted to PRIDE over the last year, as repeated identifications of the same peptides are performed. The total number of peptide identifications has increased 5.5-fold, however the number of unique peptide identifications in PRIDE has only doubled.
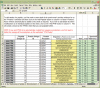
Figure 3.
A single sheet from the ProteomeHarvest PRIDE Submission Spreadsheet—Peptide Identification Data Entry.
Figure 4.
The PRIDE BioMart: results summary view for a simple query. The filter and display attributes can be seen on the left.
Similar articles
- PRIDE: a public repository of protein and peptide identifications for the proteomics community.
Jones P, Côté RG, Martens L, Quinn AF, Taylor CF, Derache W, Hermjakob H, Apweiler R. Jones P, et al. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D659-63. doi: 10.1093/nar/gkj138. Nucleic Acids Res. 2006. PMID: 16381953 Free PMC article. - The PRIDE proteomics identifications database: data submission, query, and dataset comparison.
Jones P, Côté R. Jones P, et al. Methods Mol Biol. 2008;484:287-303. doi: 10.1007/978-1-59745-398-1_19. Methods Mol Biol. 2008. PMID: 18592187 - PRIDE: Data submission and analysis.
Vizcaíno JA, Reisinger F, Côté R, Martens L. Vizcaíno JA, et al. Curr Protoc Protein Sci. 2010 Apr;Chapter 25:25.4.1-25.4.11. doi: 10.1002/0471140864.ps2504s60. Curr Protoc Protein Sci. 2010. PMID: 20393974 - The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013.
Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H. Vizcaíno JA, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D1063-9. doi: 10.1093/nar/gks1262. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203882 Free PMC article. - Proteomics data exchange and storage: the need for common standards and public repositories.
Jiménez RC, Vizcaíno JA. Jiménez RC, et al. Methods Mol Biol. 2013;1007:317-33. doi: 10.1007/978-1-62703-392-3_14. Methods Mol Biol. 2013. PMID: 23666733 Review.
Cited by
- Analysis of affinity purification-related proteomic data for studying protein-protein interaction networks in cells.
Kattan RE, Ayesh D, Wang W. Kattan RE, et al. Brief Bioinform. 2023 Mar 19;24(2):bbad010. doi: 10.1093/bib/bbad010. Brief Bioinform. 2023. PMID: 36682002 Free PMC article. Review. - Gene-selective transcription promotes the inhibition of tissue reparative macrophages by TNF.
Dichtl S, Sanin DE, Koss CK, Willenborg S, Petzold A, Tanzer MC, Dahl A, Kabat AM, Lindenthal L, Zeitler L, Satzinger S, Strasser A, Mann M, Roers A, Eming SA, El Kasmi KC, Pearce EJ, Murray PJ. Dichtl S, et al. Life Sci Alliance. 2022 Jan 13;5(4):e202101315. doi: 10.26508/lsa.202101315. Print 2022 Apr. Life Sci Alliance. 2022. PMID: 35027468 Free PMC article. - Phosphoproteome profiling uncovers a key role for CDKs in TNF signaling.
Tanzer MC, Bludau I, Stafford CA, Hornung V, Mann M. Tanzer MC, et al. Nat Commun. 2021 Oct 18;12(1):6053. doi: 10.1038/s41467-021-26289-6. Nat Commun. 2021. PMID: 34663829 Free PMC article. - An expanded mouse testis transcriptome and mass spectrometry defines novel proteins.
Gamble J, Chick J, Seltzer K, Graber JH, Gygi S, Braun RE, Snyder EM. Gamble J, et al. Reproduction. 2020 Jan;159(1):15-26. doi: 10.1530/REP-19-0092. Reproduction. 2020. PMID: 31677600 Free PMC article. - The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics.
Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, MacCoss MJ. Pino LK, et al. Mass Spectrom Rev. 2020 May;39(3):229-244. doi: 10.1002/mas.21540. Epub 2017 Jul 9. Mass Spectrom Rev. 2020. PMID: 28691345 Free PMC article. Review.
References
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. Pride: the proteomics identifications database. Proteomics. 2005;5:3537–3545. - PubMed
- Mead JA, Shadforth IP, Bessant C. Public proteomic ms repositories and pipelines: available tools and biological applications. Proteomics. 2007;7:2769–2786. - PubMed
- Editorial. Democratizing proteomics data. Nat. Biotechnol. 2007;25:262. - PubMed
- Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 2006;5:1843–1849. - PubMed