Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer - PubMed (original) (raw)
Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer
Matthew R Whorton et al. J Biol Chem. 2008.
Abstract
G protein-coupled receptors (GPCRs) are seven transmembrane domain proteins that transduce extracellular signals across the plasma membrane and couple to the heterotrimeric family of G proteins. Like most intrinsic membrane proteins, GPCRs are capable of oligomerization, the function of which has only been established for a few different receptor systems. One challenge in understanding the function of oligomers relates to the inability to separate monomeric and oligomeric receptor complexes in membrane environments. Here we report the reconstitution of bovine rhodopsin, a GPCR expressed in the retina, into an apolipoprotein A-I phospholipid particle, derived from high density lipoprotein (HDL). We demonstrate that rhodopsin, when incorporated into these 10 nm reconstituted HDL (rHDL) particles, is monomeric and functional. Rhodopsin.rHDL maintains the appropriate spectral properties with respect to photoactivation and formation of the active form, metarhodopsin II. Additionally, the kinetics of metarhodopsin II decay is similar between rhodopsin in native membranes and rhodopsin in rHDL particles. Photoactivation of monomeric rhodopsin.rHDL also results in the rapid activation of transducin, at a rate that is comparable with that found in native rod outer segments and 20-fold faster than rhodopsin in detergent micelles. These data suggest that monomeric rhodopsin is the minimal functional unit in G protein activation and that oligomerization is not absolutely required for this process.
Figures
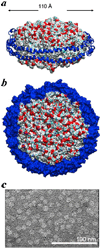
FIGURE 1. Illustration of reconstituted HDL particles
a and b, molecular models illustrating a dimer of apoA-I proteins wrapped around a phospholipid bilayer consisting of 160 POPC molecules. Each apoA-I protein (blue) is depicted as a ribbon diagram. c, electron micrograph of rHDL particles reveal a monodisperse and homogeneous rHDL particles. Images in a and b were produced using PyMol (DeLano Scientific LLC, Palo Alto, CA). Coordinates for the HDL model from Segrest et al. (60) were obtained on line and used with permission from Dr. Stephen Harvey.
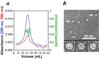
FIGURE 2. Rhodopsin incorporated in rHDL and resolved by size exclusion chromatography is photoactivable
a, purified bovine rhodopsin incorporated into rHDL particles was resolved on a Superdex 200 size exclusion column; fractions (200 µl) were collected and analyzed for λ = 280 nm (blue) and λ = 500 nm (red) absorbance. b, peak fraction from the gel filtration run was adjusted to pH 6.5, and UV-visible spectra were measured before and after photoactivation.
FIGURE 3. Rhodopsin exists as a monomer in rHDL particles
a, rhodopsin incorporated into rHDL particles was purified on a ConA-Sepharose column and subsequently resolved on a size exclusion column. Eluates were assayed for absorbance at λ = 280 nm (blue) and λ = 500 nm (red), and the rhodopsin: apoA-I ratio (green) was calculated as described under “Experimental Procedures.” b, TEM of negatively stained purified rhodopsin·rHDL particles showing their homogeneous discoidal shapes and monodispersed distribution.
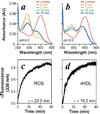
FIGURE 4. Meta II formation and decay
UV-visible spectra were measured from a rhodopsin·rHDL preparation before (red), and at 0 (orange), 5 (olive), 15 (green), and 30 min (blue) after photoactivation at pH 6.5 (a) or pH8.0 (b). The rate of meta II decay was measured by exciting 10 n
m
rhodopsin in ROS membranes (c) or rHDL (Rho·rHDL) (d) at λ = 295 nm and measuring emission at λ = 330 nm emission following a 15-s photoactivation using the methods of Farrens and Khorana (20). The relaxation times (τ) shown were calculated from three independent experiments.
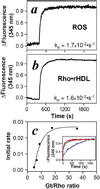
FIGURE 5. Activation of transducin by monomeric rhodopsin in rHDL
Photoactivated oligomeric rhodopsin in ROS membranes (a) or monomeric rhodopsin in rHDL (b) activates transducin at identical rates. Transducin was added to pre-activated photoactivated rhodopsin in rHDL or in ROS membranes at a 9:1 ratio as described under “Experimental Procedures.” Transducin activation was measured by monitoring the tryptophan fluorescence (λem = 345 nm) of the Gtα subunit. Initial rates based on the least squares fit of the data to an exponential activation are illustrated in the lower corner of each panel and were calculated from three independent experiments. c, elevating the Gt:rhodopsin ratio increased the rate of Gt activation (initial rate, s−1) in rhodopsin·rHDL particles (Rho·rHDL). The effect of increasing Gt:rhodopsin ratios appears to be a titration of the observed rates. The maximal rate was achieved at a Gt:rhodopsin ratio of 12:1.
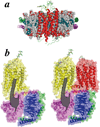
FIGURE 6. Only one receptor in a receptor dimer is necessary to activate G proteins
a, molecular model of a rhodopsin monomer in a rHDL (cross-section, side view). b, model illustrating monomeric rhodopsin in comparison to oligomeric (dimeric) rhodopsin coupling to a single G protein heterotrimer. The activated form of rhodopsin (meta II, yellow) in a dimeric form with rhodopsin (red) in a complex with transducin: Gtα (magenta), Gβ1 (blue), and Gγ1 (green). The coordinates for rhodopsin (61) and transducin (62) were obtained from the RCSB Protein Data Bank, accession number 1F88 and 1GOT, respectively. The conceptual model of a dimeric rhodopsin, where one molecule is activated, with a single Gt was previously proposed by Filipek et al. (52).
Similar articles
- Phospholipids are needed for the proper formation, stability, and function of the photoactivated rhodopsin-transducin complex.
Jastrzebska B, Goc A, Golczak M, Palczewski K. Jastrzebska B, et al. Biochemistry. 2009 Jun 16;48(23):5159-70. doi: 10.1021/bi900284x. Biochemistry. 2009. PMID: 19413332 Free PMC article. - Phosphatidylethanolamine enhances rhodopsin photoactivation and transducin binding in a solid supported lipid bilayer as determined using plasmon-waveguide resonance spectroscopy.
Alves ID, Salgado GF, Salamon Z, Brown MF, Tollin G, Hruby VJ. Alves ID, et al. Biophys J. 2005 Jan;88(1):198-210. doi: 10.1529/biophysj.104.046722. Epub 2004 Oct 22. Biophys J. 2005. PMID: 15501933 Free PMC article. - Role of membrane integrity on G protein-coupled receptors: Rhodopsin stability and function.
Jastrzebska B, Debinski A, Filipek S, Palczewski K. Jastrzebska B, et al. Prog Lipid Res. 2011 Jul;50(3):267-77. doi: 10.1016/j.plipres.2011.03.002. Epub 2011 Mar 22. Prog Lipid Res. 2011. PMID: 21435354 Free PMC article. Review. - Relevance of rhodopsin studies for GPCR activation.
Deupi X. Deupi X. Biochim Biophys Acta. 2014 May;1837(5):674-82. doi: 10.1016/j.bbabio.2013.09.002. Epub 2013 Sep 13. Biochim Biophys Acta. 2014. PMID: 24041646 Review.
Cited by
- The complex nature of CXCR4 mutations in WHIM syndrome.
Rodríguez-Frade JM, González-Granado LI, Santiago CA, Mellado M. Rodríguez-Frade JM, et al. Front Immunol. 2024 Jul 5;15:1406532. doi: 10.3389/fimmu.2024.1406532. eCollection 2024. Front Immunol. 2024. PMID: 39035006 Free PMC article. Review. - Mutant dominant-negative rhodopsin ∆I256 causes protein aggregates degraded via ERAD and prevents normal rhodopsin from proper membrane trafficking.
Cao B, Dahlen JV, Sen M, Beyer T, Leonhard T, Kilger E, Arango-Gonzalez B, Ueffing M. Cao B, et al. Front Mol Biosci. 2024 May 17;11:1369000. doi: 10.3389/fmolb.2024.1369000. eCollection 2024. Front Mol Biosci. 2024. PMID: 38828393 Free PMC article. - Investigating the Role of Rhodopsin F45L Mutation in Mouse Rod Photoreceptor Signaling and Survival.
Poria D, Kolesnikov AV, Lee TJ, Salom D, Palczewski K, Kefalov VJ. Poria D, et al. eNeuro. 2023 Mar 7;10(3):ENEURO.0330-22.2023. doi: 10.1523/ENEURO.0330-22.2023. Print 2023 Mar. eNeuro. 2023. PMID: 36823167 Free PMC article. - Computationally designed GPCR quaternary structures bias signaling pathway activation.
Paradis JS, Feng X, Murat B, Jefferson RE, Sokrat B, Szpakowska M, Hogue M, Bergkamp ND, Heydenreich FM, Smit MJ, Chevigné A, Bouvier M, Barth P. Paradis JS, et al. Nat Commun. 2022 Nov 11;13(1):6826. doi: 10.1038/s41467-022-34382-7. Nat Commun. 2022. PMID: 36369272 Free PMC article. - A quantitative ex vivo study of the interactions between reconstituted high-density lipoproteins and human leukocytes.
Pedersbæk D, Jønsson K, Madsen DV, Weller S, Bohn AB, Andresen TL, Simonsen JB. Pedersbæk D, et al. RSC Adv. 2020 Jan 23;10(7):3884-3894. doi: 10.1039/c9ra08203d. eCollection 2020 Jan 22. RSC Adv. 2020. PMID: 35492676 Free PMC article.
References
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR. Science. 2001;291:1304–1351. - PubMed
- Gilman AG. Annu. Rev. Biochem. 1987;56:615–649. - PubMed
- Angers S, Salahpour A, Bouvier M. Annu. Rev. Pharmacol. Toxicol. 2002;42:409–435. - PubMed
- Ramon E, Marron J, del Valle L, Bosch L, Andres A, Manyosa J, Garriga P. Vision Res. 2003;43:3055–3061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- EY08061/EY/NEI NIH HHS/United States
- R01 EY008061/EY/NEI NIH HHS/United States
- R01 GM079191/GM/NIGMS NIH HHS/United States
- P60 DK020572/DK/NIDDK NIH HHS/United States
- R01 EY008061-22/EY/NEI NIH HHS/United States
- GM068603/GM/NIGMS NIH HHS/United States
- GM07919/GM/NIGMS NIH HHS/United States
- R01 GM068603/GM/NIGMS NIH HHS/United States
- P60DK-20572/DK/NIDDK NIH HHS/United States
- R01 GM079191-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources