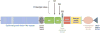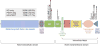Notch signaling in leukemia - PubMed (original) (raw)
Review
Notch signaling in leukemia
Jon C Aster et al. Annu Rev Pathol. 2008.
Abstract
Recent discoveries indicate that gain-of-function mutations in the Notch1 receptor are very common in human T cell acute lymphoblastic leukemia/lymphoma. This review discusses what these mutations have taught us about normal and pathophysiologic Notch1 signaling, and how these insights may lead to new targeted therapies for patients with this aggressive form of cancer.
Figures
Figure 1
Domain organization of human Notch1. Epidermal growth factor–like repeats 11–12, implicated in binding Jagged- and Delta-like ligands, are highlighted as hatched blue rectangles. The sites of cleavage by furin-like proteases (S1), ADAM-type metalloproteases (S2), and γ-secretase (S3) are shown. HDN and HDC, N- and C-terminal portions of the heterodimerization domain.
Figure 2
Molecular steps involved in canonical Notch signaling. The three key steps—ADAM- type-metalloprotease (MP) cleavage (1), γ-secretase cleavage (2), and nuclear translocation of intracellular Notch (ICN) (3)—are highlighted. Other events and involved molecules are described in the text. DSL, Delta/Serrate/Lag-2-type Notch ligand; MAML, Mastermind-like coactivator; Fringe, Fringe family glycosyltransferases; CSL, CBF1, Suppressor of Hairless, Lag-1 transcription factor.
Figure 3
The structural basis for recruitment of Mastermind-like 1 (MAML1) to binary complexes of the ankyrin repeat domain of Notch1 (ANK) and the transcription factor CSL. The MAML1 polypeptide binds as a kinked helix to a composite interface created by the association of ANK with CSL. The MAML1 helix is rendered as a green helical ribbon. MAML1 side chains that approach within 4 Å of ANK or CSL are shown as sticks. The surface of ANK is colored dark purple where an atom of MAML1 approaches within 4 Å and colored light purple elsewhere. The surface of CSL is colored dark orange where an atom of MAML1 approaches within 4 Å and colored light orange elsewhere. MAML1 makes alternating contacts with both ANK (platforms I and II) and CSL, the latter via interactions with C- and N-terminal Rel homology regions. Figure adapted with permission from Reference .
Figure 4
Distribution of Notch1 mutations in human T cell acute lymphoblastic leukemia/lymphoma (T-ALL). Each symbol represents the position of a mutation found in T-ALL cell lines (black triangles) or a series of 96 primary T-ALL samples (white triangles). The percentages of tumors with various combinations of mutations are also given. HDN and HDC, N- and C-terminal portions of the heterodimerization domain. Figure adapted with permission from Reference .
Figure 5
Activating leukemia-associated mutations within the Notch1 heterodimerization domain fall into two distinct mechanistic types. Class 1 mutations either result in subunit dissociation (Type IA, red) or lead to increased instability of the heterodimerization domain (Type IB, blue). Type II mutations (green), which are typically insertions of 12–15 residues, result in increased ligand-independent proteolytic sensitivity without detectably affecting heterodimerization domain stability. The positions of mutations of each type are shown overlaid onto a schematic representation of Notch1.
Figure 6
Structure of the Notch2 negative regulatory region (NRR) and sites corresponding to heterodimer domain (HD) mutations of Notch1 found in T cell acute lymphoblastic leukemia/lymphoma (T-ALL). (a) Two views of the Notch2 NRR, related by a 90° rotation, are shown. In the ribbon representation of the NRR, the Lin12/Notch repeat (LNR) modules are colored two shades of purple and the HD is colored in two shades of cyan; the light and dark cyans represent residues N and C terminal to the furin cleavage loop, respectively. Three bound Ca2+ ions are green, a bound Zn2+ ion is blue, and the 10 disulfide bonds are red. The positions of S1 and S2 cleavage are indicated with red arrows. (b) Sites of tumor-associated Notch1 HD mutations mapped onto the Notch2 NRR structure. (Left) The LNRs are shown in light purple and the HD backbone as a light cyan ribbon. Hydrophobic core residues are shown in ball and stick form, and side chains of residues corresponding to tumor-associated mutations are colored blue. (Right) Enlargement of the HD region, denoting the Notch2 NRR residues corresponding to known leukemia-associated HD mutations found in Notch1. Figure adapted with permission from Reference .
Figure 7
Candidate intracellular Notch1 (ICN1) negative regulatory motifs. Conserved serine and threonine residues lying in the C termini of human Notch1 and Drosophila Notch are colored. A motif containing four consecutive S residues that influence T cell acute lymphoblastic leukemia/lymphoma (T-ALL) development in mouse models (2522SSSS2525) is shown in green. The blue box indicates a site that is targeted by Fbw7; blue residues are sites of phosphorylation by cyclin-dependent kinase 8. The arrow depicts the position of the most C-terminal mutation of Notch1 yet found in T-ALL.
Figure 8
Conserved residues in the ankyrin repeat domain of Notch (ANK) mediate ANK-ANK interactions in the crystal lattice. The structure of two copies of the ANK/CSL/Mastermind-like 1/DNA complex related by crystallographic symmetry is shown. The protein subunits are rendered as ribbons (ANK, purple; CSL, gold; and Mastermind-like 1, green) and the DNA is rendered as blue sticks. Residues of ANK engaged in lattice contacts are shown as sticks in cyan. The cartoon below the structure depicts the head-to-head orientation of the two CSL binding sites in the DNA. Figure adapted with permission from Reference .
Figure 9
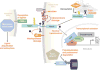
Reverse phase protein microarray profiling of Notch signals in T cell acute lymphoblastic leukemia/lymphoma (T-ALL) cell lines. Lysates were prepared from six γ-secretase inhibitor (GSI)-sensitive and six GSI-resistant T-ALL cell lines treated with 1 μM compound E, a potent GSI, or vehicle (dimethylsulfoxide) for seven days. After spotting on slides, changes in epitope levels were determined directly by incubation with Alexa Fluor 647 conjugated antibodies. Relative amounts of epitopes of interest were determined by fluorescence signal intensity and subjected to statistical analysis of microarrays. In the image shown, blue and yellow depict epitopes decreased and increased by GSI treatment, respectively. A single asterisk (*) denotes components of the mammalian target of rapamycin pathway. A double asterisk (**) identifies phospholipase D1 (PLD1), which may act upstream of mammalian target of rapamycin. Note also that p27/kip1, a cyclin-dependent kinase inhibitor, is upregulated in GSI-sensitive cell lines.
Figure 10
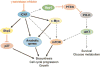
Pathways downstream of Notch1 in T cell acute lymphoblastic leukemia/lymphoma (T-ALL). Black arrows and red bars indicate proposed positive and negative regulatory interactions, respectively. PI3-K, phosphoinositol 3-kinase; mTOR, mammalian target of rapamycin; ICN1, intracellular Notch1.
Figure 11
Notch1 and T cell development. This figure depicts stages of early T cell development, which are characterized by the immunophenotypes shown below each stage. Notch1 function is required to establish the earliest identifiable intrathymic T cell progenitors (ETPs), and is also required during subsequent maturation to the DN3a stage. β-selection occurs during the DN3a to DN3b transition and is associated with dynamic changes in Notch1 and c-Myc expression. Notch1 deficiency blocks progression beyond the β-selection checkpoint, whereas c-Myc deficiency prevents only the expansion of DN3 cells. DN, CD4−/CD8− double negative; ISP, intermediate single positive; DP, CD4+/CD8+ double positive; TCR, T cell receptor.
Similar articles
- T cell acute lymphoblastic leukemia/lymphoma: a human cancer commonly associated with aberrant NOTCH1 signaling.
Pear WS, Aster JC. Pear WS, et al. Curr Opin Hematol. 2004 Nov;11(6):426-33. doi: 10.1097/01.moh.0000143965.90813.70. Curr Opin Hematol. 2004. PMID: 15548998 Review. - An activating intragenic deletion in NOTCH1 in human T-ALL.
Haydu JE, De Keersmaecker K, Duff MK, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Ferrando A. Haydu JE, et al. Blood. 2012 May 31;119(22):5211-4. doi: 10.1182/blood-2011-10-388504. Epub 2012 Apr 17. Blood. 2012. PMID: 22510873 Free PMC article. Clinical Trial. - Notch in T-ALL: new players in a complex disease.
Koch U, Radtke F. Koch U, et al. Trends Immunol. 2011 Sep;32(9):434-42. doi: 10.1016/j.it.2011.06.005. Epub 2011 Jul 19. Trends Immunol. 2011. PMID: 21775206 Review. - Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease.
Ren M, Cowell JK. Ren M, et al. Blood. 2011 Jun 23;117(25):6837-47. doi: 10.1182/blood-2010-07-295725. Epub 2011 Apr 28. Blood. 2011. PMID: 21527531 Free PMC article. - The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia.
Durinck K, Wallaert A, Van de Walle I, Van Loocke W, Volders PJ, Vanhauwaert S, Geerdens E, Benoit Y, Van Roy N, Poppe B, Soulier J, Cools J, Mestdagh P, Vandesompele J, Rondou P, Van Vlierberghe P, Taghon T, Speleman F. Durinck K, et al. Haematologica. 2014 Dec;99(12):1808-16. doi: 10.3324/haematol.2014.115683. Epub 2014 Oct 24. Haematologica. 2014. PMID: 25344525 Free PMC article.
Cited by
- Regulation of immune responses and tolerance: the microRNA perspective.
Chen CZ, Schaffert S, Fragoso R, Loh C. Chen CZ, et al. Immunol Rev. 2013 May;253(1):112-28. doi: 10.1111/imr.12060. Immunol Rev. 2013. PMID: 23550642 Free PMC article. Review. - Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Marchesini M, Gherli A, Montanaro A, Patrizi L, Sorrentino C, Pagliaro L, Rompietti C, Kitara S, Heit S, Olesen CE, Møller JV, Savi M, Bocchi L, Vilella R, Rizzi F, Baglione M, Rastelli G, Loiacono C, La Starza R, Mecucci C, Stegmaier K, Aversa F, Stilli D, Lund Winther AM, Sportoletti P, Bublitz M, Dalby-Brown W, Roti G. Marchesini M, et al. Cell Chem Biol. 2020 Jun 18;27(6):678-697.e13. doi: 10.1016/j.chembiol.2020.04.002. Epub 2020 May 7. Cell Chem Biol. 2020. PMID: 32386594 Free PMC article. - Ferroptosis and EMT: key targets for combating cancer progression and therapy resistance.
Ren Y, Mao X, Xu H, Dang Q, Weng S, Zhang Y, Chen S, Liu S, Ba Y, Zhou Z, Han X, Liu Z, Zhang G. Ren Y, et al. Cell Mol Life Sci. 2023 Aug 19;80(9):263. doi: 10.1007/s00018-023-04907-4. Cell Mol Life Sci. 2023. PMID: 37598126 Free PMC article. Review. - The telomere complex and the origin of the cancer stem cell.
Torres-Montaner A. Torres-Montaner A. Biomark Res. 2021 Nov 4;9(1):81. doi: 10.1186/s40364-021-00339-z. Biomark Res. 2021. PMID: 34736527 Free PMC article. Review. - Hyperactivation of mTORC1 and mTORC2 by multiple oncogenic events causes addiction to eIF4E-dependent mRNA translation in T-cell leukemia.
Schwarzer A, Holtmann H, Brugman M, Meyer J, Schauerte C, Zuber J, Steinemann D, Schlegelberger B, Li Z, Baum C. Schwarzer A, et al. Oncogene. 2015 Jul;34(27):3593-604. doi: 10.1038/onc.2014.290. Epub 2014 Sep 22. Oncogene. 2015. PMID: 25241901
References
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. - PubMed
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. Describes the discovery of Notch1 through analysis of the t(7;9) found in T-ALL and provides the first suggestion that truncation of Notch1 via proteolysis might play a role in its activation. - PubMed
- Aster J, Pear W, Hasserjian R, Erba H, Davi F, et al. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harb Symp Quant Biol. 1994;59:125–36. - PubMed
- Das I, Craig C, Funahashi Y, Jung KM, Kim TW, et al. Notch oncoproteins depend on γ-secretase/presenilin activity for processing and function. J Biol Chem. 2004;279:30771–80. - PubMed
- Palomero T, Barnes KC, Real PJ, Bender JL, Sulis ML, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to γ-secretase inhibitors. Leukemia. 2006;20:1279–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical