Differential chromatin looping regulates CD4 expression in immature thymocytes - PubMed (original) (raw)
Differential chromatin looping regulates CD4 expression in immature thymocytes
Huimin Jiang et al. Mol Cell Biol. 2008 Feb.
Abstract
Runx1 binds the silencer and represses CD4 transcription in immature thymocytes. In this study, using looping chromatin immunoprecipitation and chromatin conformation capture assays, we demonstrated that interactions between Runx1 and positive elongation factor b (P-TEFb) appose the silencer and enhancer in CD4-negative thymoma cells and double-negative immature thymocytes. This chromatin loop decoys P-TEFb away from the promoter, thus preventing RNA polymerase II from elongating on the CD4 gene. In the absence of Runx1 on the silencer, P-TEFb interacts with the transcription complex, forming a different chromatin loop between the enhancer and the promoter, which leads to the expression of the CD4 gene in CD4-positive hybridoma cells and double-positive thymocytes. Moreover, the knockdown of CycT1 from P-TEFb abolishes both of these chromatin loops. Finally, the selective removal and restoration of Runx1 causes rapid interchanges between these chromatin loops, which reveals the plasticity of this regulatory circuit. Thus, differential looping and decoying of P-TEFb away from the promoter mediate active repression of the CD4 gene during thymocyte development.
Figures
FIG. 1.
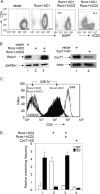
Recruitment of RNAPII, P-TEFb (CycT1), and Runx1 to the CD4 locus. (A) Schematic diagram of the CD4 locus and the amplicons used in ChIP assays. The CD4 enhancer (E) is located 12 kb upstream from the promoter (P), and the CD4 silencer (S) is located in the first intron, 2.3 kb downstream from the promoter. I designates the amplicon located between the enhancer and the promoter. (B) FACS analyses of CD4 expression in 1200M (open peak) and 3A9 (solid peak) cells stained with fluorescein isothiocyanate-conjugated anti-mouse CD4 or a rat immunoglobulin G2b(κ) isotype control (thin black line). The intensity of CD4 staining and the relative number of cells are shown on the x and y axes, respectively. (C) ChIP analyses of the CD4 locus in 1200M (CD4−) and 3A9 (CD4+) cells. Anti-RNAPII, anti-CycT1, anti-panRunx, and anti-Runx1 antibodies were used as described in Materials and Methods. Normal rabbit serum served as the negative control for antibody specificity. The presence of the CD4 enhancer, intervening sequence, promoter, and silencer in the immunoprecipitates was examined by PCR with primer sets E, I, P, and S, respectively. PCR analyses with DNA before immunoprecipitation (Input) served as controls for the amplification efficiencies of individual primer sets. PCRs were carried out at various cycle numbers to ensure linear amplification. Representative agarose gels of PCR products within the linear range are presented.
FIG. 2.
3C analyses of the CD4 locus. (A) Diagram of 3C analysis. The locations of StuI restriction sites (s) and PCR primers (arrows) used in 3C analyses, relative to the CD4 enhancer (E), promoter (P), and silencer (S), are indicated. Cross-linked and digested chromatin was ligated at very low concentrations so that only intramolecular ligation could occur. Ligation products were detected by quantitative real-time PCR with appropriate primer sets in the presence of Sybr green. The relative cross-linking frequency of the CD4 enhancer with the silencer and that of the enhancer with the promoter were evaluated by the PCR signals obtained from the ES and EP amplicons, both normalized to Hprt1 and the control template as described in Materials and Methods. (B) Negative controls of 3C analysis. Real-time PCRs were performed from StuI-digested, non-cross-linked 1200M genomic DNA and cross-linked 1200M chromatin before and after ligation. Sybr green signals from the EP and ES amplicons were normalized to signals from a fragment that was not cut by StuI. Since no signal was obtained from non-cross-linked or nonligated samples after 40 cycles (lanes 1 to 3 and 5 to 7), the signal from the EP amplicon from cross-linked and ligated 1200M chromatin (lane 8) was set to 1. (C) 3C analyses of the CD4 locus in 1200M and 3A9 cells. The y axis shows the relative cross-linking frequency of the enhancer with the promoter (EP) and of the enhancer with the silencer (ES) compared to the Hprt1 locus (set to 1; not shown on the graph). At least two independent experiments were performed for each cell population, and more than three measurements were carried out for each experiment. Results of one typical experiment are presented. Error bars, standard deviations. (D) 3C analyses of the CD4 locus in primary DN and DP thymocytes were carried out as described for panel C.
FIG. 3.
Runx1 and CycT1 knockdowns disrupt the looping between the CD4 enhancer and silencer in 1200M cells. (A) FACS analyses of CD4 expression in 1200M cells transduced either with the empty vector, with a retroviral vector containing a shRNAmir against Runx1 followed by IRES-green fluorescent protein (GFP) (Runx1-KD1), or with Runx1-KD1 plus MSCV vectors containing Runx1-IRES-human CD2 (Runx1-hCD2). The intensity of CD4 staining and the levels of enhanced GFP (EGFP) or hCD2 are presented on the y and x axes, respectively. (B) Western blotting of Runx1 and CycT1 levels in 1200M cells transduced either with the empty vector, with a retroviral vector containing a different shRNAmir against Runx1 followed by IRES-GFP (Runx1-KD2), with Runx1-KD2 plus Runx1-hCD2, or with a shRNAmir against CycT1 (CycT1-KD). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin were used as controls for equal loading. (C) FACS analyses of CD4 expression in 1200M (white peak to the left), 3A9 (dotted line to the right), Runx1-KD2 (black peak), and Runx1-KD2/Runx1-hCD2 (solid gray peak) cells stained with phycoerythrin-conjugated anti-mouse CD4. The intensity of CD4 staining and the relative numbers of cells are shown on the x and y axes, respectively. (D) 3C analyses of the CD4 locus. The relative cross-linking frequencies of the enhancer with the promoter (EP) and of the enhancer with the silencer (ES) are presented as described for Fig. 2C.
FIG. 4.
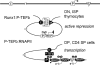
A model for active repression by Runx1. In DN and ISP thymocytes, the interaction between Runx1 and P-TEFb brings the enhancer (E) into the close proximity of the silencer (S) and prevents P-TEFb from activating RNAPII, which is arrested at the promoter (P), thus actively repressing transcription elongation. In DP and CD4 SP thymocytes, the lack of Runx1 binding to the CD4 silencer frees P-TEFb to interact with RNAPII and to activate transcription elongation. Only RNAPII on the CD4 promoter is depicted in the model.
Similar articles
- Runx1 binds positive transcription elongation factor b and represses transcriptional elongation by RNA polymerase II: possible mechanism of CD4 silencing.
Jiang H, Zhang F, Kurosu T, Peterlin BM. Jiang H, et al. Mol Cell Biol. 2005 Dec;25(24):10675-83. doi: 10.1128/MCB.25.24.10675-10683.2005. Mol Cell Biol. 2005. PMID: 16314494 Free PMC article. - Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex.
Wan M, Zhang J, Lai D, Jani A, Prestone-Hurlburt P, Zhao L, Ramachandran A, Schnitzler GR, Chi T. Wan M, et al. Eur J Immunol. 2009 Feb;39(2):580-8. doi: 10.1002/eji.200838909. Eur J Immunol. 2009. PMID: 19180471 Free PMC article. - Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes.
Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Telfer JC, et al. J Immunol. 2004 Apr 1;172(7):4359-70. doi: 10.4049/jimmunol.172.7.4359. J Immunol. 2004. PMID: 15034051 - Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.
C Quaresma AJ, Bugai A, Barboric M. C Quaresma AJ, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review. - Megakaryocytic programming by a transcriptional regulatory loop: A circle connecting RUNX1, GATA-1, and P-TEFb.
Goldfarb AN. Goldfarb AN. J Cell Biochem. 2009 Jun 1;107(3):377-82. doi: 10.1002/jcb.22142. J Cell Biochem. 2009. PMID: 19350569 Free PMC article. Review.
Cited by
- SilenceREIN: seeking silencers on anchors of chromatin loops by deep graph neural networks.
Pan JH, Du PF. Pan JH, et al. Brief Bioinform. 2023 Nov 22;25(1):bbad494. doi: 10.1093/bib/bbad494. Brief Bioinform. 2023. PMID: 38168841 Free PMC article. - Short tandem repeats are important contributors to silencer elements in T cells.
Hussain S, Sadouni N, van Essen D, Dao LTM, Ferré Q, Charbonnier G, Torres M, Gallardo F, Lecellier CH, Sexton T, Saccani S, Spicuglia S. Hussain S, et al. Nucleic Acids Res. 2023 Jun 9;51(10):4845-4866. doi: 10.1093/nar/gkad187. Nucleic Acids Res. 2023. PMID: 36929452 Free PMC article. - An autonomous TCR signal-sensing switch influences CD4/CD8 lineage choice in mice.
Basu J, Zha J, Nicolas E, Coulton M, Czyzewicz P, Hua X, Ge L, Kappes DJ. Basu J, et al. Commun Biol. 2022 Jan 21;5(1):84. doi: 10.1038/s42003-022-02999-5. Commun Biol. 2022. PMID: 35064205 Free PMC article. - ZNF382 controls mouse neuropathic pain via silencer-based epigenetic inhibition of Cxcl13 in DRG neurons.
Ma L, Yu L, Jiang BC, Wang J, Guo X, Huang Y, Ren J, Sun N, Gao DS, Ding H, Lu J, Zhou H, Zou L, Gao Y, Wang L, Sun K, Ming Y, Meng Z, Tao YX, Yan M. Ma L, et al. J Exp Med. 2021 Dec 6;218(12):e20210920. doi: 10.1084/jem.20210920. Epub 2021 Nov 11. J Exp Med. 2021. PMID: 34762123 Free PMC article. - Alprazolam Prompts HIV-1 Transcriptional Reactivation and Enhances CTL Response Through RUNX1 Inhibition and STAT5 Activation.
Lin A, Elbezanti WO, Schirling A, Ahmed A, Van Duyne R, Cocklin S, Klase Z. Lin A, et al. Front Neurol. 2021 Jul 22;12:663793. doi: 10.3389/fneur.2021.663793. eCollection 2021. Front Neurol. 2021. PMID: 34367046 Free PMC article.
References
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8327-337. - PubMed
- Bosselut, R. 2004. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat. Rev. Immunol. 4529-540. - PubMed
- de Bruijn, M. F., and N. A. Speck. 2004. Core-binding factors in hematopoiesis and immune function. Oncogene 234238-4248. - PubMed
- Ellmeier, W., S. Sawada, and D. R. Littman. 1999. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17523-554. - PubMed
- Gomez, J. A., P. Majumder, U. M. Nagarajan, and J. M. Boss. 2005. X box-like sequences in the MHC class II region maintain regulatory function. J. Immunol. 1751030-1040. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials