Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice - PubMed (original) (raw)
. 2007 Dec;134(24):4335-45.
doi: 10.1242/dev.012047.
Iphigenie Charatsi, Clive J Joyner, Chad H Koonce, Marc Morgan, Ayesha Islam, Carol Paterson, Emily Lejsek, Sebastian J Arnold, Axel Kallies, Stephen L Nutt, Elizabeth K Bikoff
Affiliations
- PMID: 18039967
- PMCID: PMC7116377
- DOI: 10.1242/dev.012047
Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice
Elizabeth J Robertson et al. Development. 2007 Dec.
Abstract
The zinc-finger transcriptional repressor Blimp1 (Prdm1) controls gene expression patterns during differentiation of B lymphocytes and regulates epigenetic changes required for specification of primordial germ cells. Blimp1 is dynamically expressed at diverse tissue sites in the developing mouse embryo, but its functional role remains unknown because Blimp1 mutant embryos arrest at E10.5 due to placental insufficiency. To explore Blimp1 activities at later stages in the embryo proper, here we used a conditional inactivation strategy. A Blimp1-Cre transgenic strain was also exploited to generate a fate map of Blimp1-expressing cells. Blimp1 plays essential roles in multipotent progenitor cell populations in the posterior forelimb, caudal pharyngeal arches, secondary heart field and sensory vibrissae and maintains key signalling centres at these diverse tissues sites. Interestingly, embryos carrying a hypomorphic Blimp1gfp reporter allele survive to late gestation and exhibit similar, but less severe developmental abnormalities, whereas transheterozygous Blimp1(gfp/-) embryos with further reduced expression levels, display exacerbated defects. Collectively, the present experiments demonstrate that Blimp1 requirements in diverse cell types are exquisitely dose dependent.
Figures
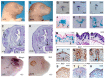
Fig. 1. Formation of the ulna and posterior forelimb digits requires Blimp1 expression.
(A-D) Alcian Blue staining of E13.5 control (+/+) and mutant (m/m) forelimbs (A,B) and hindlimbs (C,D) reveals absence of the ulna and fourth and fifth digits in Blimp1 null-embryo forelimbs, whereas hindlimbs develop normally. (E) At E16.5, Alcian Blue-Alizarian Red staining shows that growth, patterning and ossification of the residual long bones and digits proceeds normally in the mutant. (F-H) Whole-mount in situ hybridisation (WISH) showing Blimp1 expression is initially confined to the posterior mesoderm at E9.5 (F) and is rapidly downregulated over the next 48 hours (G,H). At E11.5, Blimp1 expression is also detectable in the AER. (I) WISH shows that the Blimp1-Cre transgene faithfully recapitulates the pattern of endogenous Blimp1 expression. Cell-fate mapping studies in the forelimb demonstrate that Blimp1+ cells initially confined to the posterior mesenchyme at E10.5 (J), give rise to the posterior half of the limb bud by E12.5 (K,K′). (L,L′) At E14.5, these mesodermal derivatives form the entire fourth and fifth posterior digits and the posterior half of the third digit, and the ulna. Blimp1+ cells also give rise to the posterior muscles, and connective tissues (L′). Diffuse staining in the dorsal limbs is due to the presence of /_acZ_-expressing endothelium. The fate map observed for Blimp1+ cells in the hindlimbs was identical. A, anterior; P, posterior; R, radius; U, ulna.
Fig. 2. Blimp1-deficient forelimbs fail to maintain the ZPA.
Comparison of wild-type (A,C,E,G,I,K,M,O,Q) and mutant (B,D,F,H,J,L,N,P,R) forelimb (FL) and hindlimb (HL) buds. (A-C) Activation of Shh, a marker for the ZPA, proceeds normally at E10.5 in β/_imp-_deficient limb buds, but is markedly reduced in the forelimb bud (B). By E11.5, Shh expression is confined to a small patch of distal mesenchyme in mutant forelimbs (D). (H) Blimp1 mutant limbs express Fgf4 in the forming AER at E10.5. By contrast, by E12.5 Fgf8 is absent from the mutant posterior proximal limb (J), owing to loss of the AER and underlying mesenchyme. Lmx1 expression shows normal D-V patterning in the forelimb (K,L). Tbx2 (N), and Gli1 (P) in _Blimp1_-deficient forelimb buds highlight the rapid loss of posterior tissue. Grem1 (Gremlin) is expressed in mutant limb buds albeit at lower levels (R).
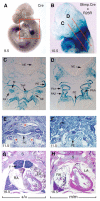
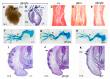
Fig. 3. Blimp1 function in progenitors of the dermal papillae is required for induction of the sensory vibrissae.
At E18.5, wild-type (A) embryos display five prominent rows of sensory vibrissae. These are completely absent from mutants (B). Frontal sections through control (C) and mutant (D) E16.5 embryos demonstrate missing vibrissae but normal tongue and tooth development. (E) Blimp1 transcripts are strongly expressed in the forming vibrissae at E12.5. (F) Shh transcription is activated but not maintained in Blimp1 mutants. (G-I) Blimp1 together with Bmp4 and Shh marks the mesenchymal condensates of the forming vibrissae at E12.5. Fate-mapping studies identify Blimp1+ cells as giving rise to the initial condensates of the prospective dermal papilla (J), and underlie the invaginating ectodermal placodes (K), ultimately giving rise to the mature DP (L). Blimp1+ cells that fail to incorporate into the DP migrate to surround the shaft of the forming follicle. (M) Fate-mapping studies show Blimp1+ cells give rise to the DP of the coat hair follicles and intrafollicular mesenchyme of the E18.5 back skin. (J,M) Punctuate lacZ staining marks endothelial cells. However in marked contrast to the vibrissae, histological analysis shows that hair follicle induction is Blimp1 independent (N,O). Ki67 staining at E12.5 shows that Blimp1 expressing cells of the dermal condensate (P), and DP (Q,R) are quiescent, whereas cells of the surface ectoderm, the ectodermally derived hair shaft and surrounding mesenchyme are strongly labelled. (S) Ki67 labelling is uniform in the mesenchyme of Blimp1 deficient embryos. (T) In wild-type embryos c-myc is detected in the surrounding mesenchyme but not in the prospective D P. (U) c-myc-positive cells are uniformly scattered throughout both the mesenchyme and surface ectoderm in sites of vibrissae induction in Blimp1 mutants. en, endothelial cells; dc, dermal condensates; dp, dermal papillae; hf, hair follicle; ifd, interfollicular demis; v, vibrissae; N, nares; T, teeth; Tg, tongue; VNO, vomeronasal organ.
Fig. 4. Blimp1 promotes expansion of the surface ectoderm and endoderm progenitors in the developing pharynx.
(A) Blimp1-Cre faithfully recapitulates the endogenous Blimp1 expression pattern at E9.5. (B) Blimp1+ cells in the pharyngeal region (boxed in A) give rise to the surface epithelium of the pharyngeal arches and the anterior heart tube. (C,D) _lacZ_-marked Blimp1+ cells contribute to both the surface ectoderm and pharyngeal endoderm of the pharyngeal arches, and ventral endoderm including the thyroid primordium, but are excluded from the pharyngeal mesoderm. Blimp1 expressed in endothelial cells marks the pharyngeal arteries. (E,F) At E11.5, the Blimp1 mutant pharynx (F) shows severe hypoplasia of the pharyngeal endoderm (PE) and pharyngeal arteries and blood vessels (asterisks) are largely absent. Frontal sections of control (G) and mutant (H) embryos show loss of the thymus in the mutants. SE, surface ectoderm; M, arch mesenchyme; NE, neurectoderm; PA pharyngeal arch; PAA, pharyngeal arch artery; PE, pharyngeal endoderm; ThP, thyroid primordium; S, sternum; T, thymus; RA, right atrium; LA left atrium.
Fig. 5
Late-onset cardiac defects in Blimp1 mutant embryos. (A-D) Transiently expressed Blimp1 in the splanchnic mesoderm anterior to the heart marks progenitors of the SHF. Blimp1+ cells contribute to the myocardium of the right ventricle, the outflow tract, and to the leaflets of the ventricular valves and walls of the aorta and pulmonary artery (D). (E-H) Outflow tract morphogenesis defects in _Blimp1_-deficient embryos (G,H) leads to persistent truncus arteriosus (PTA), in association with ventricular septal defects (VSD). Fgf8 (I-J′) and Tbx1 (K-L′) expression domains are severely downregulated, in part reflecting loss of the caudal pharyngeal arches (PA), and reduced staining in the surface epithelial and associated mesoderm. Red arrows indicate expression of Fgf8 and Tbx1 in the SHF in wild-type embryos. LV, left ventricle; RV, right ventricle; LA, left atrium; RA, right atrium; Ao, aorta; PA, pulmonary artery; V V, ventricular valves; PA, pharyngeal arch; SE, surface ectoderm; M, arch mesenchyme; PE, pharyngeal endoderm.
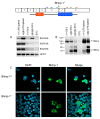
Fig. 6. Molecular characterisation of the Blimp1 gfp locus.
(Top) An IRES-gfp reporter cassette inserted 3′ to exon 6 predominantly leads to expression of truncated Blimp1 protein (Blimp-1T) lacking the C-terminal zinc fingers (Z1-Z5) owing to the presence of inframe stop codons. Arrows indicate position of PCR primers used to amplify exon 6 (blue), exon 6-8 (green) and exon 8 (red) sequences. (A) RT-PCR analysis reveals correctly spliced transcripts containing exons 6,7 and 8. (B) Western blot experiments also demonstrate wild-type protein expression in homozygous Blimp1 gfp/gfp embryos. These results are representative of three independent experiments analysing individually genotyped wild-type (_n_=8), gfp/+ (_n_=8) and gfp/gfp (_n_=7) embryos. Molecular size markers in KDa are on the left. (C) COS cells transiently transfected with expression constructs encoding full-length (FL) or truncated (T) Blimp1 protein, were stained with monoclonal anti-Blimp1 antibody and analysed by confocal microscopy. In striking contrast to the wild-type protein, truncated Blimp1 lacking the C-terminal zinc fingers fails to enter the nucleus.
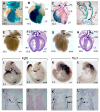
Fig. 7. Graded Blimp1 activities required for germ cell specification and development of the cardiovascular system, forelimbs and vibrissae.
(A,B) Homozygous Blimp1 gfp/gfp embryos display cardiovascular defects including PTA and VSD. Fast Red alkaline phosphatase staining of PGCs in the dorsal hindgut of wild-type (C), gfp/+ (D) and gfp/gfp (E) E9.5 embryos. Decreased numbers of PGCs are specified in gfp/+ embryos and homozygous mutants entirely lack PGCs. (F-H) Homozygous gfp/gfp forelimbs contain a rudimentary ulna and four digits (G). Heterozygous gfp/– embryos with further reduced Blimp1 activity entirely lack the ulna and are missing the posterior two digits. (I,J) Induction of the sensory vibrissae is relatively Blimp1 independent because vibrissae induction is unperturbed (arrows), and ectodermal placode induction and invagination occurs normally in gfp/gfp homozygous embryos (J). Blimp1 expression in gfp/– embryos is sufficient for coalescence of the prospective DP cells, but these fail to induce overlying ectodermal placodes (K). DC, dermal condensates; LV, left ventricle; PTA, persistent truncus arteriosus; R, radius; R V, right ventricle; U, ulna; VSD, ventricular septal defect.
Similar articles
- Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis.
Wilm TP, Solnica-Krezel L. Wilm TP, et al. Development. 2005 Jan;132(2):393-404. doi: 10.1242/dev.01572. Development. 2005. PMID: 15623803 - The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse.
Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. Vincent SD, et al. Development. 2005 Mar;132(6):1315-25. doi: 10.1242/dev.01711. Development. 2005. PMID: 15750184 - Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation.
Morgan MA, Magnusdottir E, Kuo TC, Tunyaplin C, Harper J, Arnold SJ, Calame K, Robertson EJ, Bikoff EK. Morgan MA, et al. Mol Cell Biol. 2009 Nov;29(21):5813-27. doi: 10.1128/MCB.00670-09. Epub 2009 Sep 8. Mol Cell Biol. 2009. PMID: 19737919 Free PMC article. - Germ cell specification in mice.
Hayashi K, de Sousa Lopes SM, Surani MA. Hayashi K, et al. Science. 2007 Apr 20;316(5823):394-6. doi: 10.1126/science.1137545. Science. 2007. PMID: 17446386 Review. - Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues.
John SA, Garrett-Sinha LA. John SA, et al. Exp Cell Res. 2009 Apr 15;315(7):1077-84. doi: 10.1016/j.yexcr.2008.11.015. Epub 2008 Dec 3. Exp Cell Res. 2009. PMID: 19073176 Review.
Cited by
- NonO Is a Novel Co-factor of PRDM1 and Regulates Inflammatory Response in Monocyte Derived-Dendritic Cells.
Lee K, Jang SH, Tian H, Kim SJ. Lee K, et al. Front Immunol. 2020 Jul 10;11:1436. doi: 10.3389/fimmu.2020.01436. eCollection 2020. Front Immunol. 2020. PMID: 32765503 Free PMC article. - Blimp1/Prdm1 governs terminal differentiation of endovascular trophoblast giant cells and defines multipotent progenitors in the developing placenta.
Mould A, Morgan MA, Li L, Bikoff EK, Robertson EJ. Mould A, et al. Genes Dev. 2012 Sep 15;26(18):2063-74. doi: 10.1101/gad.199828.112. Genes Dev. 2012. PMID: 22987638 Free PMC article. - Hedgehog Signal and Genetic Disorders.
Sasai N, Toriyama M, Kondo T. Sasai N, et al. Front Genet. 2019 Nov 8;10:1103. doi: 10.3389/fgene.2019.01103. eCollection 2019. Front Genet. 2019. PMID: 31781166 Free PMC article. Review. - Blimp1 expression predicts embryonic stem cell development in vitro.
Chu LF, Surani MA, Jaenisch R, Zwaka TP. Chu LF, et al. Curr Biol. 2011 Oct 25;21(20):1759-65. doi: 10.1016/j.cub.2011.09.010. Epub 2011 Oct 13. Curr Biol. 2011. PMID: 22000107 Free PMC article.
References
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. - PubMed
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. - PubMed
- Brown CB, Wenning JM, Lu MM, Epstein DJ, Meyers EN, Epstein JA. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004;267:190–202. - PubMed
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases