Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea - PubMed (original) (raw)
Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea
Kira S Makarova et al. Biol Direct. 2007.
Abstract
Background: An evolutionary classification of genes from sequenced genomes that distinguishes between orthologs and paralogs is indispensable for genome annotation and evolutionary reconstruction. Shortly after multiple genome sequences of bacteria, archaea, and unicellular eukaryotes became available, an attempt on such a classification was implemented in Clusters of Orthologous Groups of proteins (COGs). Rapid accumulation of genome sequences creates opportunities for refining COGs but also represents a challenge because of error amplification. One of the practical strategies involves construction of refined COGs for phylogenetically compact subsets of genomes.
Results: New Archaeal Clusters of Orthologous Genes (arCOGs) were constructed for 41 archaeal genomes (13 Crenarchaeota, 27 Euryarchaeota and one Nanoarchaeon) using an improved procedure that employs a similarity tree between smaller, group-specific clusters, semi-automatically partitions orthology domains in multidomain proteins, and uses profile searches for identification of remote orthologs. The annotation of arCOGs is a consensus between three assignments based on the COGs, the CDD database, and the annotations of homologs in the NR database. The 7538 arCOGs, on average, cover approximately 88% of the genes in a genome compared to a approximately 76% coverage in COGs. The finer granularity of ortholog identification in the arCOGs is apparent from the fact that 4538 arCOGs correspond to 2362 COGs; approximately 40% of the arCOGs are new. The archaeal gene core (protein-coding genes found in all 41 genome) consists of 166 arCOGs. The arCOGs were used to reconstruct gene loss and gene gain events during archaeal evolution and gene sets of ancestral forms. The Last Archaeal Common Ancestor (LACA) is conservatively estimated to possess 996 genes compared to 1245 and 1335 genes for the last common ancestors of Crenarchaeota and Euryarchaeota, respectively. It is inferred that LACA was a chemoautotrophic hyperthermophile that, in addition to the core archaeal functions, encoded more idiosyncratic systems, e.g., the CASS systems of antivirus defense and some toxin-antitoxin systems.
Conclusion: The arCOGs provide a convenient, flexible framework for functional annotation of archaeal genomes, comparative genomics and evolutionary reconstructions. Genomic reconstructions suggest that the last common ancestor of archaea might have been (nearly) as advanced as the modern archaeal hyperthermophiles. ArCOGs and related information are available at: ftp://ftp.ncbi.nih.gov/pub/koonin/arCOGs/.
Figures
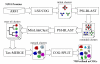
Figure 1
A flow chart of the procedure employed for the construction of the arCOGs. See Materials and Methods for the description of each step.
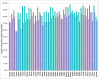
Figure 2
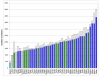
Coverage of archaeal genomes with arCOGs and COGs. Cyan, ArCOGs, purple, COGs. Abbreviations are as in Table 1.
Figure 3
Distribution of the number of species in arCOGs: three classes of archaeal genes. A semi-logarithmic plot fitted with a sum of 3 exponents
Figure 4
Distribution of phyletic patterns by the number of arCOGs. A log-log plot.
Figure 5
Functional breakdown of the entire set of arCOGs and the three core sets. EA, Euryarchaea, CA, Crenarchaea.
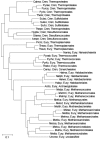
Figure 6
The gene-content tree of archaea constructed on the basis of the phyletic patterns of arCOGs. The species abbreviations are as in Table 1. Cren, Crenarchaeota; Eury, Euryarchaeota.
Figure 7
A reconstruction of gene gain and loss in archaea. Each branch is labeled by 3 numbers: black, the (inferred) number of arCOGs in the node to which the given branch leads; blue, number of arCOGs lost along the branch; red, number of arCOGs gained along the branch. The red circles on branches denote hyperthermophiles, and blue circles denote mesophiles and moderate thermophiles.
Figure 8
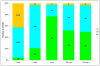
Low-bound reconstructions for ancestral archaeal forms: genomes close in size to modern hyperthermophiles. Each column shows the total number of annotated protein-coding genes in the respective archaeal species; the colored portions (green for Crenarchaeota, blue for Euryarchaeota, and cyan for Nanoarchaeota) show genes included in arCOGs. The hatched columns show the number of arCOGs assigned to LACA, the Last CrenArchaeal Common Ancestor (LCACA) and the Last EuryArchaeal Common Ancestor (LEACA).
Figure 9
Taxonomic affinities of ArCOGs with bacteria and eukaryotes. For the criteria of taxonomic assignments, see Materials and Methods.A, archaea, B, bacteria, E, eukaryotes.
Similar articles
- Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer.
Wolf YI, Makarova KS, Yutin N, Koonin EV. Wolf YI, et al. Biol Direct. 2012 Dec 14;7:46. doi: 10.1186/1745-6150-7-46. Biol Direct. 2012. PMID: 23241446 Free PMC article. - Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs).
Natale DA, Shankavaram UT, Galperin MY, Wolf YI, Aravind L, Koonin EV. Natale DA, et al. Genome Biol. 2000;1(5):RESEARCH0009. doi: 10.1186/gb-2000-1-5-research0009. Epub 2000 Nov 6. Genome Biol. 2000. PMID: 11178258 Free PMC article. - Archaeal Clusters of Orthologous Genes (arCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales.
Makarova KS, Wolf YI, Koonin EV. Makarova KS, et al. Life (Basel). 2015 Mar 10;5(1):818-40. doi: 10.3390/life5010818. Life (Basel). 2015. PMID: 25764277 Free PMC article. - A genomic perspective on protein families.
Tatusov RL, Koonin EV, Lipman DJ. Tatusov RL, et al. Science. 1997 Oct 24;278(5338):631-7. doi: 10.1126/science.278.5338.631. Science. 1997. PMID: 9381173 Review. - Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.
Koonin EV, Wolf YI. Koonin EV, et al. Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
Cited by
- The RosR transcription factor is required for gene expression dynamics in response to extreme oxidative stress in a hypersaline-adapted archaeon.
Sharma K, Gillum N, Boyd JL, Schmid A. Sharma K, et al. BMC Genomics. 2012 Jul 30;13:351. doi: 10.1186/1471-2164-13-351. BMC Genomics. 2012. PMID: 22846541 Free PMC article. - Immunoproteomic identification of immunodominant antigens independent of the time of infection in Brucella abortus 2308-challenged cattle.
Lee JJ, Simborio HL, Reyes AW, Kim DG, Hop HT, Min W, Her M, Jung SC, Yoo HS, Kim S. Lee JJ, et al. Vet Res. 2015 Mar 1;46:17. doi: 10.1186/s13567-015-0147-6. Vet Res. 2015. PMID: 25885057 Free PMC article. - Molecular evolution of the hyperthermophilic archaea of the Pyrococcus genus: analysis of adaptation to different environmental conditions.
Gunbin KV, Afonnikov DA, Kolchanov NA. Gunbin KV, et al. BMC Genomics. 2009 Dec 30;10:639. doi: 10.1186/1471-2164-10-639. BMC Genomics. 2009. PMID: 20042074 Free PMC article. - Evolution of diverse cell division and vesicle formation systems in Archaea.
Makarova KS, Yutin N, Bell SD, Koonin EV. Makarova KS, et al. Nat Rev Microbiol. 2010 Oct;8(10):731-41. doi: 10.1038/nrmicro2406. Epub 2010 Sep 6. Nat Rev Microbiol. 2010. PMID: 20818414 Free PMC article. Review. - Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia.
Hou S, Makarova KS, Saw JH, Senin P, Ly BV, Zhou Z, Ren Y, Wang J, Galperin MY, Omelchenko MV, Wolf YI, Yutin N, Koonin EV, Stott MB, Mountain BW, Crowe MA, Smirnova AV, Dunfield PF, Feng L, Wang L, Alam M. Hou S, et al. Biol Direct. 2008 Jul 1;3:26. doi: 10.1186/1745-6150-3-26. Biol Direct. 2008. PMID: 18593465 Free PMC article.
References
- Ohno S. Evolution by gene duplication. Berlin-Heidelberg-New York , Springer-Verlag; 1970.
- Galperin MY, Koonin EV. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources