Identification of novel Drosophila melanogaster microRNAs - PubMed (original) (raw)
Identification of novel Drosophila melanogaster microRNAs
Thomas Sandmann et al. PLoS One. 2007.
Abstract
MicroRNAs (miRNAs) are small non-coding RNAs with important regulatory roles in post-transcriptional regulation of metazoan development, homeostasis and disease. The full set of miRNAs is not known for any species and it is believed that many await discovery. The recent assembly of 15 insect genomes has provided the opportunity to identify novel miRNAs in the fruit fly, Drosophila melanogaster. We have performed a computational screen for novel microRNAs in Drosophila melanogaster by searching for phylogenetically conserved putative pre-miRNA structures. The ability of predicted novel miRNA precursors to be processed to produce miRNAs was experimentally verified in S2 cells and in several cases their endogenous expression at was validated by Northern blots. After experimental validation, the predictions were cross-checked with reference to a newly released set of small RNA sequences. Combining both datasets allowed us to identify 53 novel miRNA loci in the fruit fly genome 22 of which we had predicted computationally. This significantly expands the set of known miRNAs in Drosophila melanogaster. Most novel miRNAs contain unique seed sequences not found in other Drosophila miRNAs and are therefore expected to regulate novel sets of target genes. This data provides the basis for future genetic analysis of miRNA function and will aid the discovery of orthologous sequences in other species.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Example of a pre-miRNA predicted by RNAz.
A) Locus1 shows the bimodal conservation pattern typical of a conserved miRNA hairpin structure in the phastcons track of the UCSC genome browser. Sequences from Drosophilids as distant as D. mojavensis contribute to the multiz alignment at this genomic position. B) Sequences from six species were chosen as input for RNAz and lend different levels of support to stabilizing selection of the predicted secondary structure prediction (first row). The color code indicates the number of different base-pairs (green = 3 pairs to red = 1 pair) and the number of pair incompatible with the predicted structure (dark color = 0 to faint color = 2) at each position. C, D) Locus1 is predicted to fold into symmetrical hairpins in both possible directions of transcription (color code as in B).
Figure 2. Prediction of four novel miRNA clusters.
A) locus1, manual8, manual9 form a tight cluster downstream of the Grip84 locus (green). Small RNAs detected by sequencing map to each of the novel miRNA loci (matches 50–52, blue). In addition, sequences map to two loci in the vicinity, giving rise to identical mature miRNAs (matches 48,49). Finally, a sixth locus is detected immediately downstream of Grip84 (match 53). B) mir-318, located downstream of the Irp1B locus, clusters together with novel locus41 (green and small RNA sequence match 28, blue) C) Five novel miRNAs were predicted (green) and validated (blue) on either side of an exon of the CG31646 locus. D) Novel locus4 and manual5 (green) give rise to mature miRNAs (blue) and form cluster4.
Figure 3. Examples of small RNA sequences mapping to predicted miRNA loci.
Small RNA sequences recovered by sequencing were mapped to the predicted loci using Megablast. A) locus12 is transcribed from the ‘–’ strand and multiple overlapping sequences map to its coordinates. Numbers on the left indicate how often each sequence was identified in the small RNA libraries. The most abundant species (red box) most likely represents the mature miRNA. B) locus3 maps to the ‘+’ strand. Details for all predicted loci is available as supplemental data.
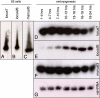
Figure 4. Northern blot validation of three miRNAs not found in small RNA libraries.
Three predicted miRNAs were not represented in the small RNA libraries, yet could be detected by Northern blot analysis. A–C) Endogenous small RNAs were detected with probes directed against locus7, locus25 and manual3 in total RNA extracted from untransfected S2 cells. D–F) All three transcripts were also detected in RNA from different embryonic stages (time after egg-laying). F) A probe against valine tRNA was used as a loading control.
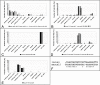
Figure 5. miRNAs sharing seed sequences.
A) The novel miRNA manual7 contains the same seed sequence as the D. melanogaster mir-12 miRNA and its orthologs. B) Sequence similarity between the novel locus11 and vertebrate miR-22 extends beyond the seed region, hinting at a possible common ancestry of these miRNAs. C) The mature sequence of Drosophila mir-12 was detected more than 1400 times in the small RNA libraries in samples from all developmental stages/tissues with the exception of the early embryo. D) The most abundant sequence mapping to the novel manual7 locus is distributed differently over the 10 sequenced samples, with RNA from discs contributing >80% of hits.
Figure 6. Four novel clusters encoding tightly co-expressed miRNAs.
Though the individual number of sequence hits recovered in the small RNA libraries differs greatly between different miRNAs, the relative abundance in the different sequenced samples is highly correlated for miRNAs originating from the same cluster. A) Similar fractions of the four members of the Drosophila miR-310-313 cluster are recovered in embryonic, imaginal disc and adult body samples, but virtually absent from the other samples. B–E) Members of the four novel miRNA clusters are distributed equally tightly over the small RNA libraries, demonstrating tight co-regulation. F) The mature sequences of locus4 and manual5 are highly similar and map to neighboring genomic coordinates (E), suggesting a duplication event that gave rise to two independent loci.
Similar articles
- Computational identification of Drosophila microRNA genes.
Lai EC, Tomancak P, Williams RW, Rubin GM. Lai EC, et al. Genome Biol. 2003;4(7):R42. doi: 10.1186/gb-2003-4-7-r42. Epub 2003 Jun 30. Genome Biol. 2003. PMID: 12844358 Free PMC article. - Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes.
Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Stark A, et al. Genome Res. 2007 Dec;17(12):1865-79. doi: 10.1101/gr.6593807. Epub 2007 Nov 7. Genome Res. 2007. PMID: 17989255 Free PMC article. - Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs.
Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Ruby JG, et al. Genome Res. 2007 Dec;17(12):1850-64. doi: 10.1101/gr.6597907. Epub 2007 Nov 7. Genome Res. 2007. PMID: 17989254 Free PMC article. - Managing the genome: microRNAs in Drosophila.
Gesellchen V, Boutros M. Gesellchen V, et al. Differentiation. 2004 Mar;72(2-3):74-80. doi: 10.1111/j.1432-0436.2004.07202003.x. Differentiation. 2004. PMID: 15066187 Review. - Resources and Methods for the Analysis of MicroRNA Function in Drosophila.
Mukherjee S, Sokol N. Mukherjee S, et al. Methods Mol Biol. 2022;2540:79-92. doi: 10.1007/978-1-0716-2541-5_3. Methods Mol Biol. 2022. PMID: 35980573 Review.
Cited by
- Evolutionary flux of canonical microRNAs and mirtrons in Drosophila.
Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. Berezikov E, et al. Nat Genet. 2010 Jan;42(1):6-9; author reply 9-10. doi: 10.1038/ng0110-6. Nat Genet. 2010. PMID: 20037610 Free PMC article. - Identification and characterization of novel conserved RNA structures in Drosophila.
Kirsch R, Seemann SE, Ruzzo WL, Cohen SM, Stadler PF, Gorodkin J. Kirsch R, et al. BMC Genomics. 2018 Dec 11;19(1):899. doi: 10.1186/s12864-018-5234-4. BMC Genomics. 2018. PMID: 30537930 Free PMC article. - Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans.
Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC. Chung WJ, et al. Genome Res. 2011 Feb;21(2):286-300. doi: 10.1101/gr.113050.110. Epub 2010 Dec 22. Genome Res. 2011. PMID: 21177960 Free PMC article. - The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation.
Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. Ye W, et al. PLoS One. 2008 Mar 5;3(3):e1719. doi: 10.1371/journal.pone.0001719. PLoS One. 2008. PMID: 18320040 Free PMC article. - Dinucleotide controlled null models for comparative RNA gene prediction.
Gesell T, Washietl S. Gesell T, et al. BMC Bioinformatics. 2008 May 27;9:248. doi: 10.1186/1471-2105-9-248. BMC Bioinformatics. 2008. PMID: 18505553 Free PMC article.
References
- Lee R, Feinbaum R, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. - PubMed
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases