Differential targeting of nuclear pore complex proteins in poliovirus-infected cells - PubMed (original) (raw)
Differential targeting of nuclear pore complex proteins in poliovirus-infected cells
Nogi Park et al. J Virol. 2008 Feb.
Abstract
Poliovirus disrupts nucleocytoplasmic trafficking and results in the cleavage of two nuclear pore complex (NPC) proteins, Nup153 and Nup62. The NPC is a 125-MDa complex composed of multiple copies of 30 different proteins. Here we have extended the analysis of the NPC in infected cells by examining the status of Nup98, an interferon-induced NPC protein with a major role in mRNA export. Our results indicate that Nup98 is targeted for cleavage after infection but that this occurs much more rapidly than it does for Nup153 and Nup62. In addition, we find that cleavage of these NPC proteins displays differential sensitivity to the viral RNA synthesis inhibitor guanidine hydrochloride. Inhibition of nuclear import and relocalization of host nuclear proteins to the cytoplasm were only apparent at later times after infection when all three nucleoporins (Nups) were cleaved. Surprisingly, analysis of the distribution of mRNA in infected cells revealed that proteolysis of Nup98 did not result in an inhibition of mRNA export. Cleavage of Nup98 could be reconstituted by the addition of purified rhinovirus type 2 2A(pro) to whole-cell lysates prepared from uninfected cells, suggesting that the 2A protease has a role in this process in vivo. These results indicate that poliovirus differentially targets subsets of NPC proteins at early and late times postinfection. In addition, targeting of interferon-inducible NPC proteins, such as Nup98, may be an additional weapon in the arsenal of poliovirus and perhaps other picornaviruses to overcome host defense mechanisms.
Figures
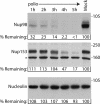
FIG. 1.
Nup98 is targeted for cleavage in infected cells. Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for the indicated length of time were analyzed by immunoblotting with monoclonal antibody 414 to detect Nup153 or monoclonal antibody MS3 to detect nucleolin or a polyclonal antisera to Nup98. All antibodies were used at 1:5,000 dilutions, and the exposure times were 10 min, 1 min, and 10 s for the Nup98, Nup153, and nucleolin immunoblots, respectively. For the percent remaining values, band intensities were quantitated by densitometry, and the amount relative to mock-infected cells are indicated. *, Cross-reactive band that is often detected with this antibody. The positions of molecular mass markers are indicated in kilodaltons.
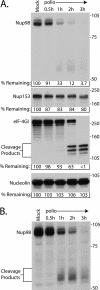
FIG. 2.
Nup98 is cleaved very rapidly after infection. (A) Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for the indicated lengths of time were analyzed by immunoblotting with polyclonal antisera to Nup98 or eIF4GI or monoclonal antibodies to detect Nup153 or nucleolin. The positions of the N-terminal eIF4GI cleavage products and molecular mass markers are indicated in kilodaltons. All antibodies were used at 1:5,000 dilutions, and the exposure times were 5 min for Nup98 and 10 s for Nup153, eIF4GI, and nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated. (B) A 30-min exposure of the immunoblot shown in panel A, with the putative Nup98 cleavage products indicated.
FIG. 3.
Effect of guanidine on cleavage of NPC proteins. Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for 2 or 5 h were analyzed by immunoblotting with the indicated antibodies. “Guanidine” refers to cells treated with 2 mM GuHCl. “Cycloheximide” refers to cells treated with 100 μg of cycloheximide/ml. Mock-infected cells treated with cycloheximide or guanidine were exposed to the reagent for 5 h. All antibodies were used at 1:5,000 dilutions, and exposure times were 1 min for Nup98, Nup153, and Nup62 and 10 s for eIF4GI and nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated.
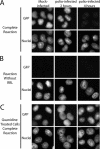
FIG. 4.
Cell-free nuclear import assays. (A) Uninfected cells (mock infected) or cells that had been infected with poliovirus for the indicated amount of time were permeabilized and used in an in vitro nuclear import assay. Assays were carried out in the presence of rabbit reticulocyte lysate (RRL) as a source of cytosolic factors. The top panels show GFP using an FITC filter, and the bottom panels show Hoechst staining of DNA using a UV filter. (B) Same as in panel A except rabbit reticulocyte lysate was omitted from the reactions. (C) Same as in panel A except that after virus adsorption the cells were incubated in the presence of 2 mM GuHCl. Mock-infected cells were exposed to GuHCl for 4 h. All GFP images were acquired by using identical exposure times and digital manipulations.
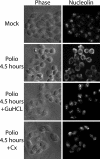
FIG. 5.
Cytoplasmic redistribution of host nuclear proteins. Uninfected cells (mock) or HeLa cells infected with poliovirus for 4.5 h were fixed and stained with an antibody directed against nucleolin. The left-hand panels show phase-contrast images of the cells (phase), while the right-hand panels show nucleolin staining of the same field (nucleolin). +GuHCl, cells treated with 2 mM GuHCl. Cx, cells treated with 100 μg of cycloheximide/ml. GuHCl and cycloheximide were added 1 h after virus adsorption.
FIG. 6.
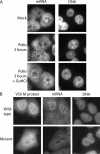
Intracellular distribution of mRNA in poliovirus-infected cells. (A) Uninfected cells (mock) or cells that had been infected with poliovirus for 3 h were analyzed by fluorescence in situ hybridization using a FITC-conjugated oligo(dT) probe. The left-hand panels show oligo(dT) staining of mRNA using an FITC filter. The right-hand panels show Hoechst staining of DNA using a UV filter. +GuHCl, infections carried out in the presence of GuHCl. (B) HeLa cells were transfected with expression vectors encoding GFP fused to wild-type VSV M protein (top panels) or the M51R mutant (bottom panels), followed by in situ hybridization using an oligo(dT) probe conjugated to Alexa Fluor 555. The GFP distribution is shown using an FITC filter, mRNA distribution is shown using a TRITC (tetramethyl rhodamine isothiocyanate) filter, and DNA is shown using a UV filter.
FIG. 7.
In vitro cleavage reactions using purified rhinovirus type 2 2Apro. (A) Uninfected HeLa whole-cell lysates were incubated in the presence or absence of the indicated amount of purified 2A protease for 4 h. Cleavage buffer refers to lysates incubated in 2Apro reaction buffer. After incubation, lysates were analyzed by immunoblotting with the indicated antibodies. Nup98, eIF4GI, and nucleolin antibodies were used at 1:2,500, 1:3,000, and 1:5,000, respectively. Exposure times were 2 min for Nup98, 10 min for eIF4GI, and 30 s for nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated. (B) Uninfected cell lysates were incubated with or without 0.1 μg of purified 2A protease for the indicated times and analyzed by immunoblotting as described above. The exposure times were 10 min for Nup98 and eIF4GI and 1 min for nucleolin. *, Putative Nup98 cleavage products. The positions of molecular mass markers are indicated in kilodaltons.
Similar articles
- RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage.
Castelló A, Izquierdo JM, Welnowska E, Carrasco L. Castelló A, et al. J Cell Sci. 2009 Oct 15;122(Pt 20):3799-809. doi: 10.1242/jcs.055988. Epub 2009 Sep 29. J Cell Sci. 2009. PMID: 19789179 - Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease.
Park N, Skern T, Gustin KE. Park N, et al. J Biol Chem. 2010 Sep 10;285(37):28796-805. doi: 10.1074/jbc.M110.143404. Epub 2010 Jul 9. J Biol Chem. 2010. PMID: 20622012 Free PMC article. - Selective Removal of FG Repeat Domains from the Nuclear Pore Complex by Enterovirus 2A(pro).
Park N, Schweers NJ, Gustin KE. Park N, et al. J Virol. 2015 Nov;89(21):11069-79. doi: 10.1128/JVI.00956-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311873 Free PMC article. - Into the basket and beyond: the journey of mRNA through the nuclear pore complex.
Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH. Ashkenazy-Titelman A, et al. Biochem J. 2020 Jan 17;477(1):23-44. doi: 10.1042/BCJ20190132. Biochem J. 2020. PMID: 31913454 Review. - Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153.
Ball JR, Ullman KS. Ball JR, et al. Chromosoma. 2005 Nov;114(5):319-30. doi: 10.1007/s00412-005-0019-3. Epub 2005 Nov 12. Chromosoma. 2005. PMID: 16133350 Review.
Cited by
- FUS/TLS Suppresses Enterovirus Replication and Promotes Antiviral Innate Immune Responses.
Xue YC, Ng CS, Mohamud Y, Fung G, Liu H, Bahreyni A, Zhang J, Luo H. Xue YC, et al. J Virol. 2021 May 24;95(12):e00304-21. doi: 10.1128/JVI.00304-21. Print 2021 May 24. J Virol. 2021. PMID: 33827951 Free PMC article. - Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity.
Schmidt HB, Görlich D. Schmidt HB, et al. Elife. 2015 Jan 6;4:e04251. doi: 10.7554/eLife.04251. Elife. 2015. PMID: 25562883 Free PMC article. - Viral subversion of the nuclear pore complex.
Le Sage V, Mouland AJ. Le Sage V, et al. Viruses. 2013 Aug 16;5(8):2019-42. doi: 10.3390/v5082019. Viruses. 2013. PMID: 23959328 Free PMC article. Review. - Structural Biology of the Enterovirus Replication-Linked 5'-Cloverleaf RNA and Associated Virus Proteins.
Pascal SM, Garimella R, Warden MS, Ponniah K. Pascal SM, et al. Microbiol Mol Biol Rev. 2020 Mar 18;84(2):e00062-19. doi: 10.1128/MMBR.00062-19. Print 2020 May 20. Microbiol Mol Biol Rev. 2020. PMID: 32188627 Free PMC article. Review. - Characterization of the African Swine Fever Virus Decapping Enzyme during Infection.
Quintas A, Pérez-Núñez D, Sánchez EG, Nogal ML, Hentze MW, Castelló A, Revilla Y. Quintas A, et al. J Virol. 2017 Nov 30;91(24):e00990-17. doi: 10.1128/JVI.00990-17. Print 2017 Dec 15. J Virol. 2017. PMID: 29021398 Free PMC article.
References
- Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, N. T. Raikhlin, L. I. Romanova, E. A. Smirnova, and E. A. Tolskaya. 1998. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252343-353. - PubMed
- Aldabe, R., E. Feduchi, I. Novoa, and L. Carrasco. 1995. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 215928-936. - PubMed
- Bablanian, R., H. J. Eggers, and I. Tamm. 1965. Studies on the mechanism of poliovirus-induced cell damage. I. The relation between poliovirus-induced metabolic and morphological alterations in cultured cells. Virology 26100-113. - PubMed
- Back, S. H., S. Shin, and S. K. Jang. 2002. Polypyrimidine tract-binding proteins are cleaved by caspase-3 during apoptosis. J. Biol. Chem. 27727200-27209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI 25105/AI/NIAID NIH HHS/United States
- R56 AI064432/AI/NIAID NIH HHS/United States
- R56 AI059467/AI/NIAID NIH HHS/United States
- P 17988/FWF_/Austrian Science Fund FWF/Austria
- P20 RR016454/RR/NCRR NIH HHS/United States
- P20 RR015587/RR/NCRR NIH HHS/United States
- AI 059467/AI/NIAID NIH HHS/United States
- P20 RR15587/RR/NCRR NIH HHS/United States
- AI 064432/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources