Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms - PubMed (original) (raw)
Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms
Michael J Moore et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Although great progress has been made in clarifying deep-level angiosperm relationships, several early nodes in the angiosperm branch of the Tree of Life have proved difficult to resolve. Perhaps the last great question remaining in basal angiosperm phylogeny involves the branching order among the five major clades of mesangiosperms (Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots). Previous analyses have found no consistent support for relationships among these clades. In an effort to resolve these relationships, we performed phylogenetic analyses of 61 plastid genes ( approximately 42,000 bp) for 45 taxa, including members of all major basal angiosperm lineages. We also report the complete plastid genome sequence of Ceratophyllum demersum. Parsimony analyses of combined and partitioned data sets varied in the placement of several taxa, particularly Ceratophyllum, whereas maximum-likelihood (ML) trees were more topologically stable. Total evidence ML analyses recovered a clade of Chloranthaceae + magnoliids as sister to a well supported clade of monocots + (Ceratophyllum + eudicots). ML bootstrap and Bayesian support values for these relationships were generally high, although approximately unbiased topology tests could not reject several alternative topologies. The extremely short branches separating these five lineages imply a rapid diversification estimated to have occurred between 143.8 +/- 4.8 and 140.3 +/- 4.8 Mya.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
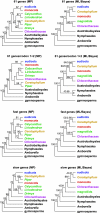
Fig. 1.
Comparison of simplified tree topologies among different partitions and phylogenetic optimization criteria. Numbers associated with branches in MP trees are MP BS support values >50%, whereas numbers associated with branches in ML trees are ML BS support values >50%/Bayesian posterior probabilities >0.5. Codon 1 + 2 refers to the combined first and second codon positions of the 61-gene combined analyses. The asterisk at the node uniting Amborella and Nymphaeales in the 61 genes/codons 1 and 2 ML/Bayes tree indicates that the GARLI ML BS analysis weakly favors a topology where Amborella is sister to all remaining angiosperms (BS = 54%).
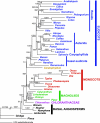
Fig. 2.
Phylogram of the best ML tree as determined by GARLI (−ln L = 460,654.151) for the 61-gene combined data set. Numbers associated with branches are ML BS values >50%/Bayesian posterior probabilities >0.5. The number in parentheses is the branch length separating outgroups from angiosperms.
Similar articles
- Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae).
Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK. Hansen DR, et al. Mol Phylogenet Evol. 2007 Nov;45(2):547-63. doi: 10.1016/j.ympev.2007.06.004. Epub 2007 Jun 16. Mol Phylogenet Evol. 2007. PMID: 17644003 - Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns.
Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL. Jansen RK, et al. Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19369-74. doi: 10.1073/pnas.0709121104. Epub 2007 Nov 28. Proc Natl Acad Sci U S A. 2007. PMID: 18048330 Free PMC article. - Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots.
Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Moore MJ, et al. Proc Natl Acad Sci U S A. 2010 Mar 9;107(10):4623-8. doi: 10.1073/pnas.0907801107. Epub 2010 Feb 22. Proc Natl Acad Sci U S A. 2010. PMID: 20176954 Free PMC article. - Nuclear phylogenomics of angiosperms and insights into their relationships and evolution.
Zhang G, Ma H. Zhang G, et al. J Integr Plant Biol. 2024 Mar;66(3):546-578. doi: 10.1111/jipb.13609. Epub 2024 Jan 30. J Integr Plant Biol. 2024. PMID: 38289011 Review. - Genome-scale data, angiosperm relationships, and "ending incongruence": a cautionary tale in phylogenetics.
Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu YL, Chase MW, Farris JS, Stefanović S, Rice DW, Palmer JD, Soltis PS. Soltis DE, et al. Trends Plant Sci. 2004 Oct;9(10):477-83. doi: 10.1016/j.tplants.2004.08.008. Trends Plant Sci. 2004. PMID: 15465682 Review.
Cited by
- Comparative case study of evolutionary insights and floral complexity in key early-diverging eudicot Ranunculales models.
Sharma B, Pandher MK, Alcaraz Echeveste AQ, Bravo M, Romo RK, Ramirez SC. Sharma B, et al. Front Plant Sci. 2024 Oct 30;15:1486301. doi: 10.3389/fpls.2024.1486301. eCollection 2024. Front Plant Sci. 2024. PMID: 39539296 Free PMC article. Review. - Comparative Plastomics of Plantains (Plantago, Plantaginaceae) as a Tool for the Development of Species-Specific DNA Barcodes.
Mehmood F, Li M, Bertolli A, Prosser F, Varotto C. Mehmood F, et al. Plants (Basel). 2024 Sep 25;13(19):2691. doi: 10.3390/plants13192691. Plants (Basel). 2024. PMID: 39409561 Free PMC article. - TFIIS is required for reproductive development and thermal adaptation in barley.
Ahmad I, Kis A, Verma R, Szádeczky-Kardoss I, Szaker HM, Pettkó-Szandtner A, Silhavy D, Havelda Z, Csorba T. Ahmad I, et al. Plant Cell Rep. 2024 Oct 10;43(11):260. doi: 10.1007/s00299-024-03345-1. Plant Cell Rep. 2024. PMID: 39390135 Free PMC article. - Delphinium as a model for development and evolution of complex zygomorphic flowers.
Sharma B, Pandher MK, Alcaraz Echeveste AQ, Romo RK, Bravo M. Sharma B, et al. Front Plant Sci. 2024 Aug 19;15:1453951. doi: 10.3389/fpls.2024.1453951. eCollection 2024. Front Plant Sci. 2024. PMID: 39224845 Free PMC article. Review. - The evolutionary adaptation of wood-decay macrofungi to host gymnosperms differs from that to host angiosperms.
Zhang X, Dong Y, Li Y, Wu X, Chen S, Wang M, Li Y, Ge Z, Zhang M, Mao L. Zhang X, et al. Ecol Evol. 2024 Jul 17;14(7):e70019. doi: 10.1002/ece3.70019. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39026950 Free PMC article.
References
- Soltis PS, Soltis DE. Am J Bot. 2004;91:1614–1626. - PubMed
- Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer; 2005.
- Angiosperm Phylogeny Group. Bot J Linn Soc. 2003;141:399–436.
- Mathews S, Donoghue MJ. Science. 1999;286:947–950. - PubMed
- Parkinson CL, Adams KL, Palmer JD. Curr Biol. 1999;9:1485–1488. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases