Posttranscriptional control of neuronal development by microRNA networks - PubMed (original) (raw)
Review
Posttranscriptional control of neuronal development by microRNA networks
Fen-Biao Gao. Trends Neurosci. 2008 Jan.
Abstract
The proper development of the nervous system requires precise spatial and temporal control of gene expression at both the transcriptional and translational levels. In different experimental model systems, microRNAs (miRNAs) - a class of small, endogenous, noncoding RNAs that control the translation and stability of many mRNAs - are emerging as important regulators of various aspects of neuronal development. Further dissection of the in vivo physiological functions of individual miRNAs promises to offer novel mechanistic insights into the gene regulatory networks that ensure the precise assembly of a functional nervous system.
Figures
Figure 1
The roles of miRNAs in the specification of SOPs. In non-SOP cells in the proneural cluster, enhanced Notch signaling leads to the association between Su(H) and Notch intracellular domain (NIntra), which in turn activates the transcription of E(spl). E(spl) suppresses the expression of Sens and proneural genes. To ensure a low level of Sens expression in non-SOP cells, miR-9a suppresses Sens through its 3′ UTR. In SOPs, the lack of Notch signaling leads to the formation of a repressor complex containing Su(H), which inhibits E(spl) expression. Sens expression is high and maintains proneural gene expression that endows the SOP fate. The absence of miR-9a in SOPs is partially responsible for the high level of Sens expression. miR-7 and other miRNAs may be involved in the suppression of E(spl).
Figure 2
Schematic representation of the double negative feedback loops between miRNAs and transcription factors. (a) In ASEL sensory neurons in C. elegans, a high level of lsy-6 suppresses Cog-1, which controls the expression of miR-273. (b) In ASER, a high level of miR-273 suppresses Die-1, a transcription factor required for lsy-6 expression.
Figure 3
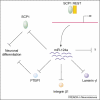
The role of miR-124a in neuronal development and its regulation by REST. The upregulation of miR-124a expression during neuronal differentiation requires the derepression by the REST-SCP1 complex. As the most abundant miRNA in the brain, miR-124a regulates the expression of many target mRNAs. Yet, the developmental consequences of lack of miR-124a in vivo remain to be further examined.
Figure 4
A negative feedback loop between an miRNA and a transcription factor in Drosophila. (a) In progenitor cells in the Drosophila eye, high-level expression of the transcription factor Yan suppresses miR-7 expression. (b) During photoreceptor differentiation, transient activation of the epidermal growth factor receptor (EGFR) signaling pathway leads to the degradation of Yan and the expression of miR-7, which further downregulates the level of Yan through binding to its 3′ UTRs.
Similar articles
- Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells.
Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Suárez Y, et al. Circ Res. 2007 Apr 27;100(8):1164-73. doi: 10.1161/01.RES.0000265065.26744.17. Epub 2007 Mar 22. Circ Res. 2007. PMID: 17379831 - Involvement of FMRP in Primary MicroRNA Processing via Enhancing Drosha Translation.
Wan RP, Zhou LT, Yang HX, Zhou YT, Ye SH, Zhao QH, Gao MM, Liao WP, Yi YH, Long YS. Wan RP, et al. Mol Neurobiol. 2017 May;54(4):2585-2594. doi: 10.1007/s12035-016-9855-9. Epub 2016 Mar 19. Mol Neurobiol. 2017. PMID: 26993298 - Decoding the ubiquitous role of microRNAs in neurogenesis.
Nampoothiri SS, Rajanikant GK. Nampoothiri SS, et al. Mol Neurobiol. 2017 Apr;54(3):2003-2011. doi: 10.1007/s12035-016-9797-2. Epub 2016 Feb 24. Mol Neurobiol. 2017. PMID: 26910816 Review. - Many routes to a micro RNA.
Yeo JH, Chong MM. Yeo JH, et al. IUBMB Life. 2011 Nov;63(11):972-8. doi: 10.1002/iub.524. IUBMB Life. 2011. PMID: 22031495 Review. - The Roles of Arabidopsis CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs.
Sun Z, Guo T, Liu Y, Liu Q, Fang Y. Sun Z, et al. PLoS Genet. 2015 Oct 16;11(10):e1005598. doi: 10.1371/journal.pgen.1005598. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26473486 Free PMC article.
Cited by
- Identification of β-Dystrobrevin as a Direct Target of miR-143: Involvement in Early Stages of Neural Differentiation.
Quaranta MT, Spinello I, Paolillo R, Macchia G, Boe A, Ceccarini M, Labbaye C, Macioce P. Quaranta MT, et al. PLoS One. 2016 May 25;11(5):e0156325. doi: 10.1371/journal.pone.0156325. eCollection 2016. PLoS One. 2016. PMID: 27223470 Free PMC article. - Developmental and functional expression of miRNA-stability related genes in the nervous system.
de Sousa É, Walter LT, Higa GS, Casado OA, Kihara AH. de Sousa É, et al. PLoS One. 2013 May 20;8(5):e56908. doi: 10.1371/journal.pone.0056908. Print 2013. PLoS One. 2013. PMID: 23700402 Free PMC article. - MicroRNA Expression Profiling Screen miR-3557/324-Targeted CaMK/mTOR in the Rat Striatum of Parkinson's Disease in Regular Aerobic Exercise.
Liu W, Li L, Liu S, Wang Z, Kuang H, Xia Y, Tang C, Yin D. Liu W, et al. Biomed Res Int. 2019 Jun 12;2019:7654798. doi: 10.1155/2019/7654798. eCollection 2019. Biomed Res Int. 2019. PMID: 31309116 Free PMC article. - The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons.
Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. Parrish JZ, et al. Neuron. 2009 Sep 24;63(6):788-802. doi: 10.1016/j.neuron.2009.08.006. Neuron. 2009. PMID: 19778508 Free PMC article. - The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila.
Xu XL, Li Y, Wang F, Gao FB. Xu XL, et al. J Neurosci. 2008 Nov 12;28(46):11883-9. doi: 10.1523/JNEUROSCI.4114-08.2008. J Neurosci. 2008. PMID: 19005053 Free PMC article.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. - PubMed
- Lee Y, et al. Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol. 2006;71:51–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources