Evolutionary history of bacteriophages with double-stranded DNA genomes - PubMed (original) (raw)
Evolutionary history of bacteriophages with double-stranded DNA genomes
Galina Glazko et al. Biol Direct. 2007.
Abstract
Background: Reconstruction of evolutionary history of bacteriophages is a difficult problem because of fast sequence drift and lack of omnipresent genes in phage genomes. Moreover, losses and recombinational exchanges of genes are so pervasive in phages that the plausibility of phylogenetic inference in phage kingdom has been questioned.
Results: We compiled the profiles of presence and absence of 803 orthologous genes in 158 completely sequenced phages with double-stranded DNA genomes and used these gene content vectors to infer the evolutionary history of phages. There were 18 well-supported clades, mostly corresponding to accepted genera, but in some cases appearing to define new taxonomic groups. Conflicts between this phylogeny and trees constructed from sequence alignments of phage proteins were exploited to infer 294 specific acts of intergenome gene transfer.
Conclusion: A notoriously reticulate evolutionary history of fast-evolving phages can be reconstructed in considerable detail by quantitative comparative genomics.
Figures
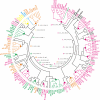
Figure 1
Bacteriophage phylogeny inferred from gene content. The tree was built using generalized average distance and neighbor-joining algorithm (see Methods). Large dots indicate clades inferred in the majority of resampled data sets. The branches leading to individual phages are colored according to their ICTV classification: family Siphoviridae is in magenta, Podoviridae is in orange, Myoviridae is in green, Fuselloviridae is in yellow, and Tectiviridae is in blue. The Bayesian tree displaying essentially the same phylogeny is presented in Fig. S1, available as Additional File 1. In the inner circle, the italicized, underlined colored numbers indicate 18 well-supported phage groups (see Supplementary Text for groups' description). They are followed by summary information of horizontal transfer events, where I stands transfer into the group, O for transfer from this group to another group, and W for transfer within the group. The algorithm for reconstructing these events is described in Methods and illustrated in Figure 2.
Figure 2
Inference of horizontal gene transfer between phages. Phage genome tree is inferred from gene content data (left side of the top panel) and sequence family trees are inferred from the aligned sequences, separately for each POG (right side of the top panel). The T-REX algorithm is used to infer HGT events by choosing such rearrangements of the gene content tree that reattach the subtrees in a way that minimizes the Robinson and Foulds topological distance to the appropriate sequence family tree. On the top right, a fragment of sequence alignment for one class of cII transcription regulators (POG226) is shown. The sequence family tree built on the basis of complete alignment is shown at the bottom right corner, and the sub-tree of the gene-content tree that contain the same set of phages as thesequence family tree is shown at the bottom left corner. Two pairs of phages, namely, 933W and Stx2I, as well as HK620 and P22, are in discordant positions in the gene content and protein family trees (indicated by the blue edges in both trees). To reconcile gene content and protein family trees, T-REX suggests a transfer from 933W to Stx2I and from HK620 to P22 (blue arrows).
Similar articles
- Reticulate representation of evolutionary and functional relationships between phage genomes.
Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Lima-Mendez G, et al. Mol Biol Evol. 2008 Apr;25(4):762-77. doi: 10.1093/molbev/msn023. Epub 2008 Jan 29. Mol Biol Evol. 2008. PMID: 18234706 - Protein repertoire of double-stranded DNA bacteriophages.
Liu J, Glazko G, Mushegian A. Liu J, et al. Virus Res. 2006 Apr;117(1):68-80. doi: 10.1016/j.virusres.2006.01.015. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16490276 Review. - A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes.
Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M. Dwivedi B, et al. BMC Evol Biol. 2013 Feb 7;13:33. doi: 10.1186/1471-2148-13-33. BMC Evol Biol. 2013. PMID: 23391036 Free PMC article. - Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site.
Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, Calendar R, Loessner MJ. Dorscht J, et al. J Bacteriol. 2009 Dec;191(23):7206-15. doi: 10.1128/JB.01041-09. Epub 2009 Sep 25. J Bacteriol. 2009. PMID: 19783628 Free PMC article. - Phage diversity, genomics and phylogeny.
Dion MB, Oechslin F, Moineau S. Dion MB, et al. Nat Rev Microbiol. 2020 Mar;18(3):125-138. doi: 10.1038/s41579-019-0311-5. Epub 2020 Feb 3. Nat Rev Microbiol. 2020. PMID: 32015529 Review.
Cited by
- New insights into the biodiversity of coliphages in the intestine of poultry.
Sørensen PE, Van Den Broeck W, Kiil K, Jasinskyte D, Moodley A, Garmyn A, Ingmer H, Butaye P. Sørensen PE, et al. Sci Rep. 2020 Sep 16;10(1):15220. doi: 10.1038/s41598-020-72177-2. Sci Rep. 2020. PMID: 32939020 Free PMC article. - Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life.
Kannan L, Li H, Rubinstein B, Mushegian A. Kannan L, et al. Biol Direct. 2013 Dec 19;8:32. doi: 10.1186/1745-6150-8-32. Biol Direct. 2013. PMID: 24354654 Free PMC article. - The proportional lack of archaeal pathogens: Do viruses/phages hold the key?
Gill EE, Brinkman FS. Gill EE, et al. Bioessays. 2011 Apr;33(4):248-54. doi: 10.1002/bies.201000091. Epub 2011 Feb 15. Bioessays. 2011. PMID: 21328413 Free PMC article. - Orthologous gene clusters and taxon signature genes for viruses of prokaryotes.
Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. Kristensen DM, et al. J Bacteriol. 2013 Mar;195(5):941-50. doi: 10.1128/JB.01801-12. Epub 2012 Dec 7. J Bacteriol. 2013. PMID: 23222723 Free PMC article. - Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation.
Grazziotin AL, Koonin EV, Kristensen DM. Grazziotin AL, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D491-D498. doi: 10.1093/nar/gkw975. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789703 Free PMC article.
References
- Virus Taxonomy . In: VIIIth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. Elsevier, Amsterdam-Boston; 2005. pp. 23–116.
- Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources